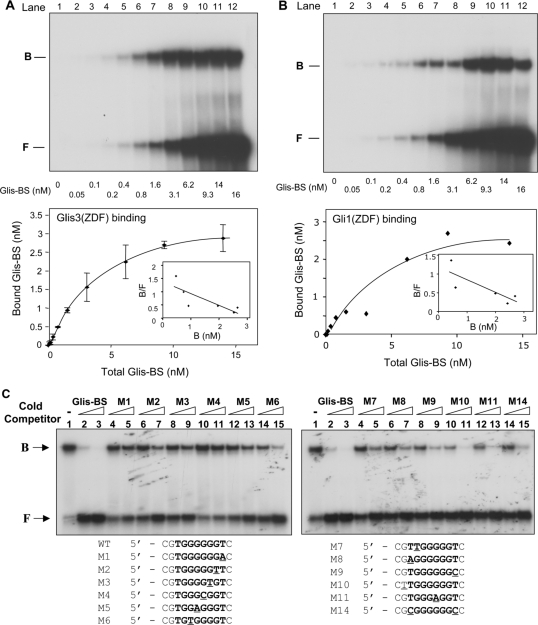
Figure 5.
Analysis of the affinity and specificity of the binding of Glis3 and Gli1 to Glis-BS. (A) Binding of Glis3 to Glis-BS. The binding of His6-Glis3(ZFD) to radiolabeled Glis-BS was examined by EMSA as a function of increasing concentrations of Glis-BS as indicated (top panel). The amount of bound and free Glis-BS was determined and the Kd calculated by Scatchard plot analysis (lower panel). (B) Binding of Gli1 to Glis-BS. The binding of His6-Gli1(ZFD) to radiolabeled Glis-BS was examined by EMSA as a function of increasing concentrations of Glis-BS as indicated (top panel). The amount of bound and free Glis-BS was determined and the Kd calculated by Scatchard plot analysis (lower panel). (C) Effect of various mutations in Glis-BS on Glis3 binding. EMSA analysis was performed with His6-Glis3(ZFD) and the 32P-labeled consensus Glis-BS (0.5 nM) in the absence and presence of 5- or 25-fold excess of unlabeled Glis-BS or mutant Glis-BS (M1-M14) as indicated. The sequences of the M1-M14 mutants are shown beneath the EMSA. The mutated nucleotides are underlined.