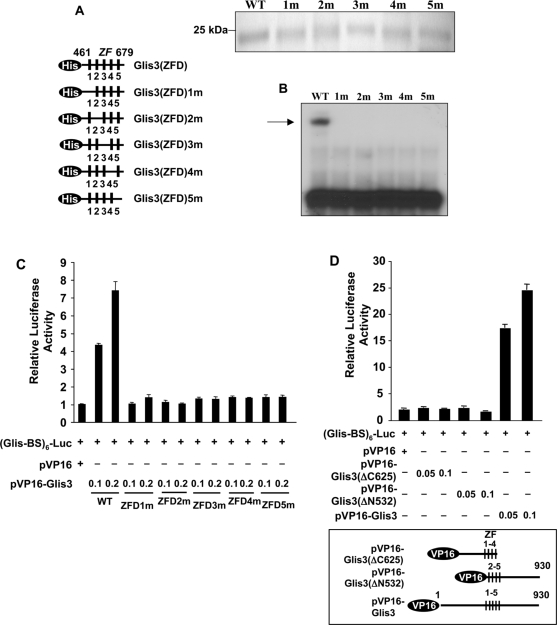
Figure 7.
All five zinc finger motifs are required for optimal binding of Glis3 to the consensus Glis-BS. (A) A schematic view of His6-Glis3(ZFD)1–5m mutants in which the first Cys in each zinc finger motif is mutated to Ala (left panel). PAGE analysis of His6-Glis3(ZFD) proteins used in EMSA (upper panel). (B) EMSA using His6-Glis3(ZFD) and His6-Glis3(ZFD)1–5m mutant proteins and 32P-labeled consensus Glis-BS. The position of the Glis3/Glis-BS complex is indicated by the arrow. (C) The ability of VP16-Glis3 and VP16-Glis3(ZFD)1–5m mutants to induce (Glis-BS)6-dependent transactivation of the LUC reporter gene was examined in HEK293T cells. (D) The ability of VP16-Glis3 and mutants VP16-Glis3(ΔC625) and VP16-Glis3(ΔN532), encoding a C- and N-terminal deletion mutant of Glis3 as indicated in the lower panel, to induce (Glis-BS)6-dependent transactivation of the LUC reporter gene was examined. The relative luciferase activity was calculated and plotted. Numbers indicate amount of plasmid (μg) transfected.