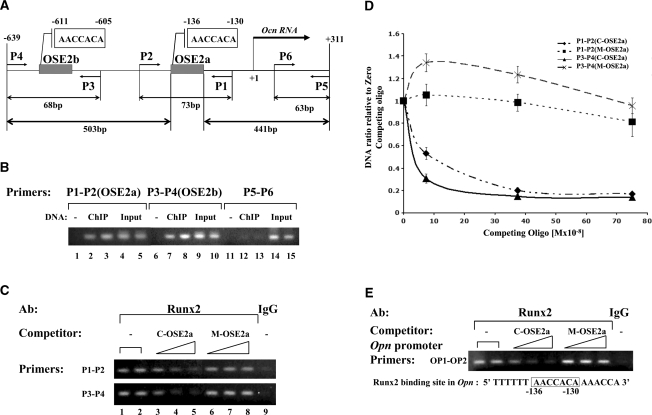
Figure 1.
Analysis of RUNX2 binding to the Ocn and Opn promoters in differentiated MC-4 cells using competitive N-ChIP. (A) Schematic representation of Runx2-binding sites in the proximal murine Ocn promoter. The positions of the RUNX2-binding sites, OSE2a and OSE2b, relative to the transcription start site are shown. P1, P2, P3, P4, P5 and P6 represent the different PCR primers used for the analysis of ChIP DNA (see Table 1). P1-P2 and P3-P4 were used to analyze the Runx2 binding to OSE2a and OSE2b, respectively. The sizes of the fragments they amplify are indicated at the bottom of the figure. (B) Primers pairs (P1, P2) and (P3, P4) specifically detect Runx2 interactions with OSE2a and OSE2b, respectively. Shown is the analysis of PCR products obtained with two independent X-ChIP-DNA samples obtained using Runx2 antibody and different primer pairs: P1-P2, P3-P4 and P5-P6 (see panel A). Control PCRs were done with duplicate samples of Input-DNAs and without DNA for each primer combination. Note that the P5-P6 primer pair amplifies a DNA fragment 441-bp away from OSE2a and that these primers do not produce a PCR product with ChIP-DNA (lanes 12 and 13). In contrast, the fragment amplified by the P3-P4 primer pair is a similar distance (503 bp) from OSE2a and, in this case the PCR is clearly positive (lanes 7 and 8). (C) N-ChIP competition assay for the Ocn promoter. The C-OSE2a double-stranded oligonucleotide or the mutant M-OSE2a was added at increasing concentrations (75, 375 and 750 nM) to immunoprecipitation reactions. A control reaction was also performed with normal rabbit IgG. The gels are ethidium bromide-stained agarose gels of the PCR products obtained with ChIP DNAs using the Ocn promoter-primers P1-P2 (top) and P3-P4 (bottom). (D) Real-time PCR analysis of the N-ChIP-competition assay. ChIP-DNAs were quantified by real-time PCR using two primer-pairs, P1-P2 and P3-P4, and their respective TaqMan probes. Results are expressed as ChIP-DNA ratio relative to zero competing-oligo from two independent experiments. The legend indicates the primer pair used in the PCR and, in parenthesis, the competing-oligo used for each ChIP-DNA set of samples. (E) N-ChIP competition assay for the Opn promoter. ChIP-DNAs were analyzed by PCR using the Opn promoter primers OP1 and OP2. These primers correspond to the region near the Runx2-binding site in the Opn promoter (−130 to −136) having the same sequence as the OSE2 site (AACCACA, lower panel).