Abstract
Secondary bile acids, formed by intestinal bacteria, are suggested to play a significant role in cancers of the gastrointestinal tract in humans. Bile acid 7α/β-dehydroxylation is carried out by a few species of intestinal clostridia which harbor a multi-gene bile acid inducible (bai) operon. Several genes encoding enzymes in this pathway have been cloned and characterized. However, no gene product(s) has yet been assigned to the production of 3-oxo-Δ4-cholenoic acid intermediates of cholic acid (CA), chenodeoxycholic acid (CDCA) or ursodeoxycholic acid (UDCA). We previously reported the baiH gene encodes an NADH:flavin oxidoreductase (NADH:FOR); however, the role of this protein in bile acid 7-dehydroxylation is unclear. Homology searches and secondary structural alignments suggest this protein to be similar to flavoproteins which reduce α/β-unsaturated carbonyl compounds. The baiH gene product was expressed in E. coli, purified and discovered to be a stereospecific NAD(H)-dependent 7β-hydroxy-3-oxo-Δ4-cholenoic acid oxidoreductase. Additionally, high sequence similarity between the baiH and baiCD gene products suggests the baiCD gene may encode a 3-oxo-Δ4-cholenoic acid oxidoreductase specific for CDCA and CA. We tested this hypothesis using cell extracts prepared from E. coli overexpressing the baiCD gene and discovered that it encodes a stereo-specific NAD(H)-dependent 7α-hydroxy-3-oxo-Δ4-cholenoic acid oxidoreductase.
Keywords: bile acid 7-dehydroxylation; flavoprotein; Old yellow enzyme; enoate reductase; 2,4-dienoyl CoA reductase; ursodeoxycholic acid; Clostridium; baiCD; baiH
1. Introduction
Bile acids are synthesized in the liver from cholesterol. Prior to secretion, bile acids are conjugated to either glycine or taurine and actively transported from the liver via canalicular transporters (1). Conjugated bile acids-also termed bile salts-are stored in the gallbladder along with unesterified cholesterol, phospholipids and conjugated bilirubin. Upon meal induced hormonal stimulation, the gallbladder contracts, emptying its contents into the small intestine. Bile salts promote the solubilization and absorption of fats, cholesterol, and fat-soluble vitamins in the small intestine. The majority of bile salts are recovered in the ileum by active transport and returned to the liver in the portal circulation (1). This process of enterohepatic circulation of bile salts occurs several times each day. During this cycling, several hundred milligrams of bile salts are lost into the human colon.
Colonic bacteria are responsible for the biotransformation of conjugates of cholic acid (CA) (3α, 7α, 12α-trihydroxy-5β-cholan-24-oic acid) and chenodeoxycholic acid (CDCA) (3α,7α-dihydroxy-5β-cholan-24-oic acid) into the unconjugated secondary bile acids deoxycholic acid (DCA) (3α,12α-dihydroxy-5β-cholan-24-oic acid) and lithocholic acid (LCA) (3α-hydroxy-5β-cholan-24-oic acid), respectively (2). Unlike rodents, the human liver lacks the ability to 7α-hydroxylate secondary bile acids. Lithocholic acid, due to its intrinsic cytotoxicity is detoxified by species-specific hydroxylation or sulfation and generally rapidly eliminated from the circulating bile acids. DCA is retained to a larger extent due to its higher solubility and can accumulate to high levels in the bile of some individuals and some species, notably the rabbit (2). Exposure of the colonic epithelium to secondary bile acids is strongly suggested by epidemiologic (3-5), animal models (6-8), and cell culture and signaling studies (9-13) to promote the colon carcinogenesis process.
Conjugated bile salts undergo deconjugation mediated by bacterial hydrolases in the distal small intestine and large intestine. Unconjugated bile acids can then be reversibly epimerized at the 3, 7 and 12 hydroxy groups (reviewed by 2, and 14). Ursodeoxycholic acid (UDCA) (3α, 7β-dihydroxy-5β-cholan-24-oic acid)-makes up a small percentage of the biliary pool in healthy individuals (<5%). UDCA is an important therapeutic agent for the treatment of cholestatic liver disease (15-18) and its administration has been suggested to prevent colon cancer (19-21). UDCA is capable of being directly 7β-dehydroxylated to LCA (22) as well as epimerized to CDCA and then 7α-dehydroxylated to LCA (23-25). However, genes involved in the 7β-dehydroxylation of UDCA have not yet been identified.
Several genes encoding enzymes in the bile acid 7α-dehydroxylating pathway from the human fecal isolate Clostridium scindens VPI 12708 have been cloned and characterized (2). The current model suggests that 7α-hydroxy (CA, CDCA) and 7β-hydroxy (UDCA) bile acids are actively transported across the bacterial membrane by an H+ dependent bile salt transporter (baiG) (26), conjugated to coenzyme A (baiB) (27), and oxidized at the 3-hydroxy group by NAD(P)(H) dependent 3α-hydroxysteroid dehydrogenase (baiA) (28). Oxidation of the 3α-hydroxy group is followed by the formation of 3-oxo-Δ4-cholenoic acid-CoA intermediates by unidentified NAD(P)(H)-dependent 3-oxo-Δ4-cholenoic acid oxidoreductase(s). At some point in the pathway, the CoA-ester is hydrolyzed or more likely the CoA moiety is transferred to incoming primary bile acids (2, 29-31). The baiF gene product has been shown to have bile acid CoA hydrolase activity (32); however, recent evidence indicates that the baiF belongs to a family of enzymes that have CoA transferase activity (2, 30). The gene encoding the bile acid 7α-dehydratase has been shown to be the baiE gene product; however, the 7β-dehydratase has not been identified, although is predicted to be encoded by the baiI, a homologue of the baiE located on the same bai operon (33). 7α/β-dehydration yields a 3-oxo-4,6-choladienoic acid intermediate which is then reduced in three sequential steps to DCA or LCA, or to the 5α epimers (A/B ring juncture in the cis configuration) allodeoxycholic acid or allolithocholic acid, respectively. Gene products catalyzing reactions in the reductive arms of the pathway have yet to be elucidated.
We have previously reported that the baiH gene product has NADH:flavin oxidoreductase activity (NADH:FOR) (34, 35). In addition, the baiCD gene product is found to share significant primary and secondary structural identity with the baiH gene product and related proteins whose structures have been elucidated. However, the role of the baiH and baiCD gene products in bile acid 7-dehydroxylation is unclear. In the current article, we provide evidence that the baiCD and baiH gene products from Clostridium scindens VPI 12708 encode stereo-specific NAD(H)-dependent 3-oxo-Δ4-cholenoic acid oxidoreductases involved in bile acid 7α-dehydroxylation and bile acid 7β-dehydroxylation, respectively.
2. Materials and methods
2.1. Bacterial strains, culture media, and conditions
Clostridium scindens VPI 12708 (formerly Eubacterium sp. strain VPI 12708) was cultivated under anaerobic conditions in Brain Heart Infusion (BHI) broth containing sodium cholate (100 μM) and maintained as previously described (36). Clostridium absonum (ATCC 27555) was purchased from ATCC. C. absonum was initially grown in cooked meat broth for 24 hrs at 37°C and subsequently stored at 4°C and used for no more than 3 weeks as starter culture source. The cells were cultured in BHI supplemented with 0.2 % fructose (w/v). E. coli BL21-CodonPlus (DE3)-RIL was purchased from Stratagene (La Jolla, CA) and used for expression of recombinant proteins. Recombinant E. coli cells were grown with shaking in modified TYGPN (tryptone 20 g, yeast extract 10 g, 0.8 % glycerol, 5 g Na3HPO4, 10 g KNO3, and 3 μM hematin per liter; pH 7.0) containing 100 μg/ml ampicillin.
2.2. Polyacrylamide gel electrophoresis and immunoblot analysis
Protein samples were separated and analyzed on a 10% SDS-polyacrylamide gel electrophoresis (PAGE) (37). Protein samples were prepared for SDS-PAGE by heating for 5 min at 100°C in an equal volume of sample buffer (0.1 M Tris-HCl, 5 % SDS, 0.9 % 2-mercaptoethanol (2ME), 20 % glycerol, pH 6.8). Gels were stained with 0.2 % (w/v) Coomassie brilliant blue R-250 (Sigma, St. Louis, MO) in ethanol: acetic acid: water (30:10:60, v/v/v). A broad-range protein weight marker (Bio-Rad, Hercules, CA) was used for protein size determinations. For western blot analysis, proteins were similarly separated by SDS-PAGE, and electrophoretically transferred to a nitrocellulose membrane by electroblotting using a Bio-Rad Immuno-Blot assay kit. Antiserum against purified baiH gene product was raised in rabbits as described previously (35). Detection of the baiH gene product on nitrocellulose membranes was performed using a goat anti-rabbit alkaline phosphatase conjugate.
2.3. PCR and recombinant DNA methods
Total genomic DNA from C. scindens VPI 12708 was purified as described previously (38) and used as a template for PCR amplification of the baiH gene using primers baiHF-PstI (5’-TAGCACTGCAGGCCTATGTT-3’) containing PstI restriction site and baiHR-SacI (5’-ATCAGAGCTCATTGCCTTCAC-3’) containing SacI restriction site. The PCR reaction mixture contained 100 ng of C. scindens VPI 12708 genomic DNA, 100 pmol each primer, 0.3 mM dNTPs, Pfu polymerase buffer, and 2.5 U Pfu DNA polymerase in a total volume of 50 μl. Cycling parameters included an initial 3 min denaturation at 94°C followed by 30 cycles of amplification 94°C for 35 s, 52°C for 30 s and 68°C for 2 min followed by one additional 7 min elongation at 68°C. PCR was performed on a DNA Thermal cycler (MJ research, Watertown, MA). The PCR product and pSport1 (Invitrogen, Carlsbad, CA) vector were double-digested with PstI and SacI, gel purified (GENECLEAN spin kit) and ligated with T4 DNA ligase (NEB). The plasmid was transformed into E. coli DH5α (Bio-line, Randolph, MA) by electroporation using an Eppendorf 2510 electroporator at 2500V. The recombinant plasmid (pSport1-baiH) was purified from E. coli DH5α and transformed into E. coli BL21-CodonPlus (DE3)-RIL for overexpression of baiH gene product. The sequence was verified at the Virginia Commonwealth University Nucleic Acid Core Facility using standard primers.
2.4. Enzyme Assays
NADH:FOR activity was determined at 25°C by monitoring the oxidation of NADH at 340 nm (ε340=6.22 mM-1cm-1) using a Beckman DU-640 spectrophotometer (Fullerton, CA). The standard assay mixture (1 ml) contained 100 mM sodium phosphate buffer (pH 6.8), 150 μM NADH, 150 μM FAD and an appropriate amount of enzyme added to initiate the reaction. Assays were performed under aerobic conditions in a 1 ml quartz cuvette with a 1 cm pathlength. One unit of NADH:FOR activity is defined as the amount of enzyme catalyzing the oxidation of 1 μmol of NADH min-1mg-1 of protein. Protein concentrations were determined according to Bradford (39) using the Bio-Rad protein assay kit, with BSA as standard for routine assays.
3-oxo-Δ4-cholenoic acid oxidoreductase activity assays were performed in reaction mixtures containing the following: 50 mM sodium phosphate buffer (pH 6.8), 1.5 mM NAD+, 225 μM [24-14C]-3-dehydro-UDCA (3-oxo-7β-hydroxy) or [24-14C]-3-dehydro-CDCA (3-oxo-7α-hydroxy) with a specific activity of 0.01 μCi μmole-1 substrate) and purified recombinant BaiH protein (100 μg). The reaction mixture (1 ml) was incubated at 37 °C for 4 hrs. 3/18/2008to ~3.0 with 1 N HCl. Radiolabeled bile acid metabolites were extracted with ethyl acetate, dried under a stream of nitrogen gas, dissolved in 100 μl MeOH, spotted, separated by thin layer chromatography on Baker flex silica gel IB-2 plates (J.T Baker Chemical Co, Philipsburg, NJ) using a solvent system of iso-octane: ethyl acetate: acetic acid (15:10:2 v/v/v) and radiolabeled bile acid metabolites were visualized by autoradiography (Kodak MS film, Rochester, N.Y). Sections of TLC silica gel containing bile acid reaction products were scraped and radioactivity quantified by liquid scintillation spectrometry (Packard, Meriden, CT). Extracts prepared from E. coli containing pSport plasmids without baiH or baiCD gene inserts served as a negative controls.
2.5. Expression and purification of recombinant baiH gene product
E. coli BL21-CodonPlus(DE3)-RIL harboring pSport1-baiH were grown in 5 ml of TYGPN medium (tryptone 20 g; yeast extract 10 g; glycerol 87 mM; sodium phosphate 35 mM, pH 7.0; KNO3 100 mM; hematin 3 μM) with 100 μg of ampicillin per ml at 37°C for 8 hrs. The culture was stored at 4°C overnight. The next day, cells were collected by centrifugation (3,000 × g, 10 min) and suspended in 5 ml of TYGPN medium containing 100 μg/ml ampicillin. This cell suspension was used to inoculate 2 liter flasks containing 600 ml TYGPN broth medium containing 100 μg/ml ampicillin. The cells were cultivated at 37°C with shaking at 250 rpm for about 3 hr to reach an A600 of about 0.4 at which point the cultures were induced by addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to 1 mM final concentration. Cultures were grown at 25°C for an additional 4 hr at which time the cells were harvested by centrifugation at 5,500 × g, 15 min at 4°C. The cells were suspended in 400 ml of buffer A (50 mM sodium phosphate buffer; pH 6.8, 2% (w/v) glycerol, 15 mM of 2-mercaptoethanol (2ME)) cells were lysed by sonicating twice for 10 min intervals and cell debris removed by centrifugation at 21,000 × g for 30 min. The supernatant fluid containing soluble recombinant BaiH was removed for further purification.
All purification methods were followed essentially as described previously with some modifications (34). Frozen cells were suspended in buffer A, disrupted by sonication on ice, and centrifuged at 21,000 × g for 30 min. Protein was concentrated and protein under 50 kDa removed using Centriprep YM50 (50 kDa cut-off) concentrators (Millipore, Billerica, MA). The protein solution was then applied to a 2 × 10 cm Protein–Pak DEAE 40HR column (Waters, Milford, MA) equilibrated with buffer A. After the column was washed with 60 ml of buffer A, NADH:FOR was eluted with a 500 ml linear gradient of NaCl (0 to 1.0 M) in buffer A at a flow rate of 2 ml min-1. Fractions with a yellow color and NADH:FOR activity were pooled and dialyzed overnight against 2 L of the same buffer. The dialyzed solution was loaded onto a 1.0 × 5.5 cm Cibacron blue 3GA agarose column (Sigma, St. Louis, MO) equilibrated with buffer A. The column was then washed with 100 ml of buffer A, and protein partially eluted with 20 ml of buffer A containing 1mM NADH, 1 mM FAD, and 20 mM dithiothreitol (DTT). The remaining protein was eluted with buffer A containing 1M KCl. Active protein fractions eluted with NAD and FAD were pooled and loaded onto a 1.0 × 6.4 cm Bio-Scale CHT5-1 hydroxyapatite column (Bio-Rad, Hercules, CA) equilibrated with buffer C (5 mM potassium phosphate buffer pH 7.0, 2% glycerol, 15 mM 2-ME). After the column was washed with buffer C at 1ml min-1, active fractions were eluted with a linear gradient of 0 to 0.5 M potassium phosphate (pH 7.0). The purified protein from this step was dialyzed with buffer A, concentrated by a Centriprep YM50 concentrator and stored in buffer A containing 50% glycerol (w/v) at -20 °C. Protein remained active for 4 weeks under these conditions.
2.6. Preparation of bile acid substrates
[24-14C]-CDCA was purchased from American Radiolabeled Chemicals, Inc. (St. Louis, Mo), and determined to be 99.9% pure by TLC. C. absonum expresses both 7α-HSDH and 7β-HSDH and can be used to efficiently epimerize CDCA to UDCA (40, 41). [24-14C]-UDCA was biologically synthesized from [24-14C]-CDCA using Clostridium absonum ATCC 27555 grown in BHI. C. absonum was induced at O.D600nm 0.4 by addition of 200 μM sodium chenodeoxycholate (CDCA) plus 2 μCi of [24-14C]-CDCA for production of [24-14C]-UDCA (0.01 μCi μmole-1). After 24 hr growth following induction, the pH of the culture medium was lowered to ~ pH 3 and bile acid metabolites were extracted twice with ethyl acetate, dried under a stream of nitrogen gas and dissolved in 100 μl of absolute methanol (MeOH). [24-14C] CDCA, [24-14C] 7-oxo-LCA and [24-14C] UDCA were separated by TLC as described above. Non-radiolabeled CDCA and UDCA standards (Sigma) were also chromatographed along with the radiolabeled reaction products and this portion of the plate stained with phosphomolybdic acid for detection. Radiolabeled bands co-migrating with UDCA standard (Sigma) were scraped and extracted with ethyl acetate, dried, and suspended in MeOH.
[24-14C] UDCA was converted to [24-14C] 3-dehydro-UDCA (3-oxo-7β-hydroxy) enzymatically using commercially available 3α-hydroxysteroid dehydrogenase (Sigma, St. Louis, Mo) in the following reaction mixture: 0.5 ml total volume 50 mM sodium phosphate buffer (pH 6.8), 1 U 3α-hydroxysteroid dehydrogenase, 1.5 mM NAD+, 100 μM unlabeled UDCA and 1 μCi [24-14C]-UDCA. The reaction mixture was allowed to incubate for 4 h at 37 °C. Bile acids were separated by TLC as described above and radiolabeled bile acid reaction products were detected by autoradiography.
2.7. Purification and mass spectrometry of bile acid products
The analysis and identification of BaiH catalyzed bile acid reaction product was carried out on a Shimadzu HPLC (Kyoto, Japan) with a SIL-HTC Autosampler by reverse-phase HPLC using a Brownlee RP-300 (Perkin Elmer, Wellesley, MA) C8 7m 2.1 i.d. × 100 mm column. The mobile phase consisted of 10 mM ammonium acetate (solvent A) and 10 mM ammonium acetate in acetonitrile (solvent B). The column was equilibrated in solvent A. After injection of a 5 ml sample of the bile acid, solvent B was increased from 0-100% at 10% min-1. The column was regenerated by switching back to solvent A and re-equilibrating for 5 min. The flow rate was 0.2 ml min-1 and the eluted material was passed into the electrospray ionization interface of a MDS Sciex/Applied Biosystems API 4000 Q Trap (Foster city, CA) operating in the negative ion mode. Mass spectra were recorded over a 350-700 m/z range.
2.8. Protein sequence analysis
Homology searches with the baiH amino acid sequence from C. scindens VPI 12708 (AAC45417) were made against the UniProt database. Multiple sequence alignments were constructed with the T-COFFEE program available on the World Wide Web (http://www.ch.embnet.org/software/TCoffee.html) (42). Clostridium scindens VPI 12708 baiH (AAC45417) and Clostridium hiranonis baiCD (AAF22846) genes were sequenced previously (43). Secondary structural predictions were carried out with the PSIPRED program (http://bioinf.cs.ucl.ac.uk/psipred/) (44, 45). Secondary structural predictions were compared with the known secondary structure from the crystal structure of 2, 4-dienoyl CoA reductase (dcr) from Escherichia coli (46).
2.9. Nucleotide sequence accession number
The baiC and baiD genes from C. scindens were originally reported as separate genes, however, after discovery that the baiCD is a single gene product in C. hiranonis, the baiC and baiD gene region was resequenced from C. scindens and found to encode a single gene (D.H. Mallonee and P.B. Hylemon, unpublished data). Also, a baiH gene has been located downstream of the bai operon in C. hiranonis and has not been reported previously (J.E. Wells and P.B. Hylemon, unpublished data). GenBank accession numbers for the baiCD gene from C. scindens VPI 12708 is EF539210 and the baiH gene from C. hiranonis TO931 is EF539211.
3. Results
3.1. Purification of recombinant BaiH
Initially, the baiH gene was cloned into a pET24a (+) vector with C-terminal hexa-histidine tag to make a BaiH-His recombinant. The BaiH-His showed NADH:FOR activity in crude extract prior to nickel affinity chromatography. However, following nickel affinity chromatography, the purified enzyme lost detectable NADH:FOR activity. Problems with His-tag use has also been reported for the YqjM protein from Bacillus subtilis; a baiH homologue (47). The quarternary structure of recombinant His-tagged YqjM was reported to be altered by the His-tag resulting in expression of an inactive enzyme. This is not the case with the baiH gene product as the enzyme showed NADH:FOR activity in crude extract similar to previously reported in recombinant BaiH lacking affinity tag (34) before His-binding chromatography (data not shown). The BaiH is known to contain 1 mol copper per subunit (34). While the mechanism for this loss of activity is not known, we speculate that the nitriloacetic acid, which chelates Ni2+, Cu2+, and Co2+, and in turn binds the hexa-histidine tail, may have chelated copper from the enzyme causing inactivation (48, 49). Addition of various concentrations of Cu2+ to the pure enzyme did not restore activity. Therefore, we opted to clone the gene into the pSport1 vector without an affinity tag.
SDS-PAGE analysis of the active fractions after CHT5-1 hydroxyapatite chromatography revealed a highly enriched protein with a subunit molecular mass of 72 kDa that was reactive against anti-BaiH antiserum (Figure 1). The specific activity and purification fold were dramatically increased by Cibacron Blue 3GA and hydroxyapatite chromatography (Table I). The baiH gene product was purified 23–fold over the crude extract. The final enzyme preparation had a NADH:FOR specific activity of 16.4 U mg-1. NADH:FOR enzyme activity existed in bright yellow fractions as observed previously (34).
Figure 1. SDS-PAGE and immunoblot analysis of baiH gene product (NADH:FOR) overexpression and purification.
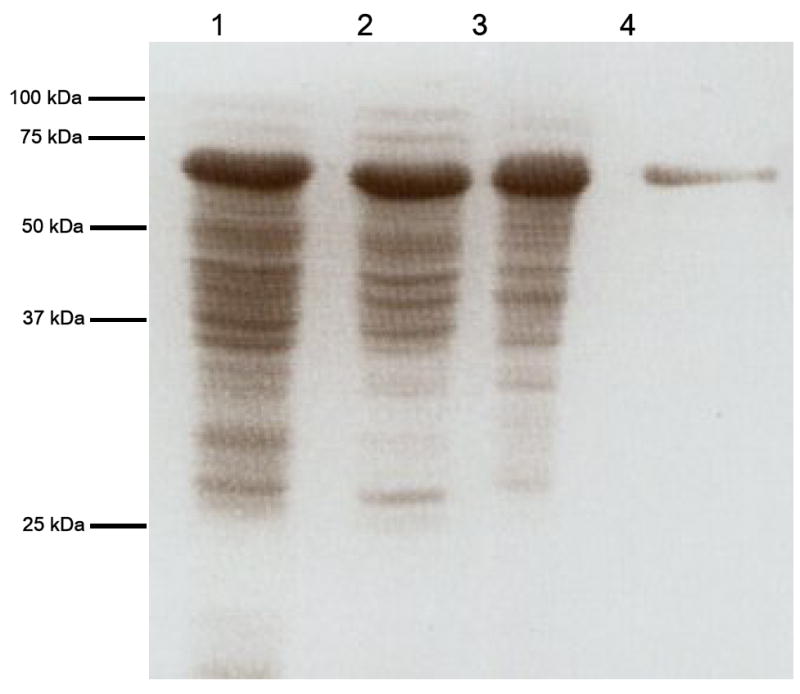
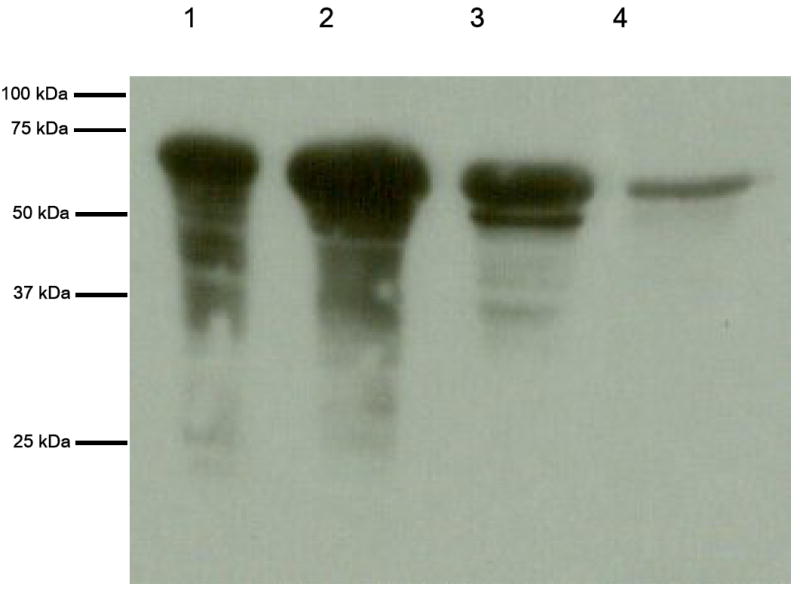
(a.) Proteins were separated on a 10 % SDS-polyacrylamide gel and stained with Coomassie Blue R-250. Lane 1, cell free extract after IPTG induction (20 μg loaded); Lane 2, flow-through from DEAE 40HR (20 μg loaded); Lane 3, flow-through from Cibacron Blue 3GA agarose (20 μg loaded); lane 4, flow-through from Hydroxyapatite CHT5-1 column (5 μg of purified enzyme loaded). (b.) Immunoblot of SDS-PAGE using anti-BaiH antiserum.
Table 1.
Purification scheme of baiH gene protein in E.coli BL21 CodonPlus(DE3)-RIL(pSportI-baiH)
| Step | Total Volume (mL) | Total Protein (mg/mL) | Total activity (U) | Specific activity (U/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|---|
| Cell free extract | 35 | 410 | 93.3 | 0.7 | 1.0 | 100 |
| Centriprep treatment | 20 | 175 | 80 | 1.2 | 1.7 | 86 |
| DEAE 40 HR Cellulose | 11 | 92 | 66 | 3 | 4.3 | 71 |
| Cibacron blue 3GA agarose | 5.2 | 63 | 55.5 | 8 | 11.4 | 60 |
| Hydroxyapatite CHT | 2.3 | 28 | 35.1 | 16.4 | 23.4 | 38 |
3.2. Stereospecific NAD(H)-dependent 3-oxo-Δ4-cholenoic acid oxidoreductase activity
We tested the hypothesis that the baiH gene product encodes an NAD(H)-dependent 3-oxo-Δ4-cholenoic acid oxidoreductase by addition of substrates [24-14C] 3-dehydro-UDCA or [24-14C] 3-dehydro-CDCA with NAD+ and purified BaiH. A single reaction product of more hydrophilic character was detected by autoradiography following TLC when [24-14C] 3-dehydro-UDCA and NAD+ were substrates (Figure 2). Previous studies have shown the specificity of this enzyme for NAD(H) (34) and the BaiH did not show 3- oxo-Δ4-cholenoic acid oxidoreductase activity with NADP(H) (data not shown). The bile acid metabolite generated following BaiH catalyzed reaction with NAD+ was scraped from TLC plates, pooled, further purified by ion exchange chromatography and identified by electrospray mass spectrometry (data not shown). The deprotonated molecular ion given by the product was 2 Da lighter than that given by the 3-dehydro-CDCA substrate, consistent with a loss of 2 hydrogens from the substrate and potentially indicating a 3-dehydro-Δ4-CDCA structure. Previous observations showed loss of the 5β hydrogen during incubation of [5β-3H] [24-14C] CA to CA induced whole cells and cell extracts of C. scindens VPI 12708 (50). Taken together, these observations strongly suggest the formation of a 3-dehydro-Δ4-UDCA structure.
Figure 2. TLC autoradiograph of BaiCD and BaiH catalyzed NAD(H)-dependent 3-oxo-Δ4-cholenoic acid oxidoreductase activity assays.
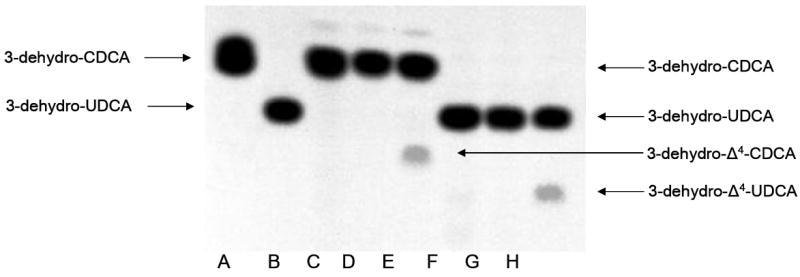
The reactions were carried out in a 500 μl volume consisting of 50 mM sodium phosphate buffer (pH 6.8) containing 225 μM of each respective substrate with a specific activity of 0.01 μCi μmole-1. A: [24-14C] 3-dehydro-CDCA standard, B: [24-14C] 3-dehydro-UDCA, C: [24-14C] 3-dehydro-CDCA + 100 μg cell-free extract of E. coli (plasmid, no insert) D: [24-14C] 3-dehydro-CDCA + 1.6 U purified BaiH, E: [24-14C] 3-dehydro-CDCA + 100 μg E.coli BL21(DE3) expressing baiCD. F: [24-14C] 3-dehydro-UDCA + 100 μg E. coli BL21(DE3) (plasmid, no insert), G: [24-14C] 3-dehydro-UDCA + 100 μg cell-free extract of E.coli BL21(DE3) expressing baiCD, H: [24-14C] 3-dehydro-UDCA + 1.6 U purified BaiH. All reactions were performed with 1.5 mM NAD+ as co-substrate.
In addition, we overexpressed the baiCD gene in E. coli BL21 (DE3) and tested for 3-oxo-Δ4-cholenoic acid oxidoreductase activity. The baiCD gene product was detected by SDS-PAGE at the expected molecular mass of 70 kDa (Figure 3). Addition of baiCD overexpressed E. coli cell extract to reaction mixture containing 1.5 mM NAD+ resulted in a product of greater hydrophilicity than the starting substrate [24-14C] 3-dehydro-CDCA (Figure 2). This product was not seen using cell-extract of E. coli BL21(DE3) containing pSport1 vector without baiCD gene insert. BaiCD did not recognize the substrate [24-14C] 3-dehydro-UDCA. The product derived from reaction with [24-14C] 3-dehydro-CDCA was scraped from TLC plates, pooled and subjected to mass-spectrometry (data not shown). The major mass ion was 2 amu less than the 3-dehydro-CDCA substrate and thus consistent with the loss of 2 hydrogens from the substrate bile acid and potentially a 3-dehydro-Δ4-CDCA structure.
Figure 3. SDS-PAGE of cell-extract of recombinant E. coli BL21(DE3) overexpressing baiCD gene.
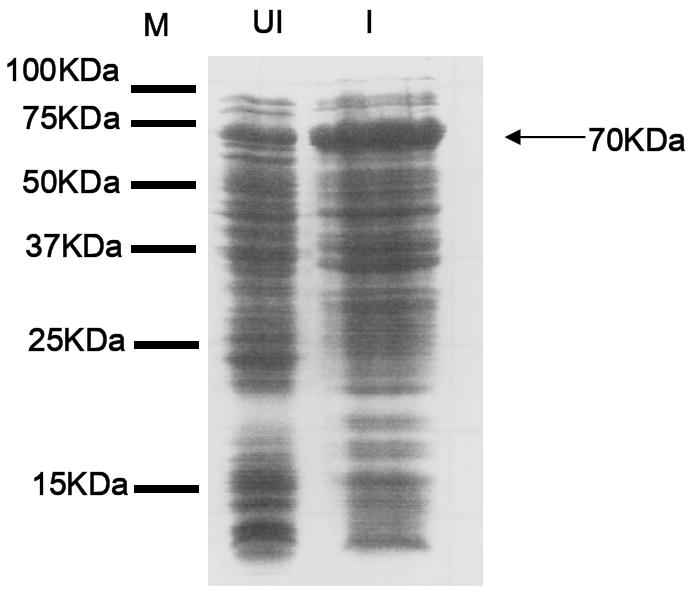
(M) Marker (UI) uninduced, (I) induced 1 mM IPTG. 30 μg protein loaded and gel stained with Coomassie brilliant blue R-250. A large band at ~ 70 kDa was detected in IPTG induced cell-extracts indicating overexpression of baiCD gene.
3.3. Amino acid sequence alignment and secondary structural prediction of the BaiH
Protein-protein BLAST searches of the baiH gene product suggests homology with a family of pyridine-nucleotide dependent flavoproteins (Table II) including NADH oxidase from Thermoanaerobacter brockii (CAA47660) (51), 2-enoate reductase from Clostridium tyrobutyricum (CAA71086) (52) and Moorella thermoacetica (CAA76082) (52), 2, 4-dienoyl-CoA reductase from several bacterial species, and Old yellow enzyme (OYE) from Thermoanaerobacter tengcongensis (NP_623692) (53). E. coli 2, 4-dienoyl CoA reductase (dcr) (AAC76116) is a 73 kDa protein which uses NADPH as an electron donor and contains a 4Fe-4S cluster, 1 mol FMN, and 1 mol FAD per mole subunit (46). The dcr gene product from E. coli shares 32 % identity and 49 % similarity at the amino acid sequence level with the baiH gene product from C. scindens VPI 12708. A 2.4 Å structure has been solved for the E. coli dcr gene product allowing a basis of comparison for secondary structure prediction of the baiH gene product (Figure 4). The predicted secondary structure of the BaiH is very similar to that of the dcr gene product suggesting a similar TIM-barrel fold. It is interesting to note the conserved cysteine consensus pattern of C-(2X)-C-(2-3X)-C-(11-12X)-C between the BaiH, E. coli Dcr and other homologues suggesting that the BaiH and BaiCD contain 4Fe-4S centers.
Table II.
Characteristics of selected BaiH homologues
| Enzyme | Organism | Subunit Mr (Kda) | Quaternary Structure | Cofacter | Electron donor | Electron Acceptor | Percent Identity/Similarity to BaiH | Accession Number | Reference |
|---|---|---|---|---|---|---|---|---|---|
| BaiH | Clostridium scindens | 72 | homotrimer | 1 FAD, Fex-Sy* | 3-dehydro-UDCA | NAD+ | 100/ 100 | AAC45417 | This study, 2, 15 |
| BaiCD | Clostridium scindens | 70 | ND | ND | 3-dehydro-CDCA | NAD+ | 32/ 46 | EF539210 | This study |
| Enoate reductase | Clostridium tyrobutyricum | 73 | homododecamer | 1 FAD, 1 FMN 1 4Fe-4S | NADH | α, β-unsaturated carbonyls | 29/ 48 | CAA71086 | 51 |
| 2,4-dienoyl CoA Reductase | Escherichia coli | 73 | monomer | 1 FAD, 1 FMN 1 4Fe-4S | NADPH | unsaturated fatty acyl-CoA | 29/ 49 | 1PS9A | 21 |
| Old Yellow Enzyme (OYE2) | Saccharomyces cerevisiae | 49.9 | homodimer | 1 FMN | NADPH | α, β unsaturated carbonyls | 23/ 42† | Q03558 | 57, 61 |
| YqjM (OYE) | Bacillus subtilis | 37.4 | homotetramer | 1 FMN | NADPH | α,β unsaturated carbonyls and nitroaromatics | 29/ 49‡ | BAA12619 | 14, 26 |
|
| |||||||||
| NoxB1 | Archaeoglobus fulgidus | 69 | monomer | FAD | NADH | unknown | 35/ 55 | NP 069291 | 25 |
Data inconclusive (2)
alignment between full length OYE2 and amino acids 1-457 of BaiH
alignment between full length YqjM and amino acids 1-351 of BaiH
Figure 4. Amino acid sequence alignment of the BaiH (AAC45417) from C. scindens VPI 12708 with protein homologues.
2,4-dienoyl-CoA reductase (dcr) (AAC76116) from E. coli, 2-enoate reductase (CAA71086) from Clostridium tyrobutyricum, the baiCD gene product from Clostridium scindens (EF539210) and Clostridium hiranonas (AAF22846) and the baiH gene product from C. hiranonis (EF539211) Secondary structural elements based on the crystal structure of DCR (21) are shown (below alignment). Predicted secondary structural elements of the BaiH protein are also shown (above alignment). Arrows correspond to beta-sheets and loops to alpha helices. Residues corresponding to FMN binding (■), FAD binding (*), and cysteine residues involved in a 4Fe-4S center (♦) from the crystal structure of DCR are also shown. The alignment was generated with the T-COFFEE program (42) and sequence similarities/identities highlighted with Boxshade 3.1 (http://www.ch.embnet.org/software/BOX_form.html).
4. Discussion
In this present study, we present evidence that the baiCD and baiH gene products encode NAD+-dependent 3-oxo-Δ4-cholenoic acid oxidoreductases recognizing 7α-hydroxy and 7β-hydroxy bile acids, respectively (Figure 5). Previously, we demonstrated loss of the 5β-hydrogen during 7α-dehydroxylation of [5β-3H] + [24-14C]-CA both in vivo and from cell extracts of CA induced C. scindens whole cells suggesting introduction of a C4-C5 double-bond (50). Addition of [24-14C] 3-oxo-Δ4-cholenoic acid to CA induced whole cells and cell-extracts of C. scindens VPI 12708 resulted in conversion to [24-14C] DCA (54). Subsequent studies elucidated the first two steps of the bile acid 7α/β-dehydroxylation pathway; CoA ligation (27) and oxidation of the 3α-hydroxy group producing a 3-dehydro-CA-CoA intermediate (28). Collectively, these data suggested the presence of an enzyme catalyzing the region-selective oxidation at C4-C5 yielding a 3-dehydro-Δ4-CA-CoA intermediate. Others have speculated that the baiH and baiCD genes were involved in C-C double bond oxidation/reduction based on homology to enzymes with similar function (55, 56). The fact that there are two homologous flavin oxidoreductases of similar Mr (70-72 kDa) with 32 % identity and 46 % similarity that are co-expressed from a bai operon suggests a possible similar function.
Figure 5. Proposed reactions catalyzed by the baiCD and baiH gene product.

R group may be the carboxylic acid or the coenzyme A conjugate
We tested this hypothesis by incubation of a crude cell extract of E. coli overexpressing the baiCD gene with [24-14C] 3-dehydro-UDCA or [24-14C] 3-dehydro-CDCA, and indeed the reaction yielded a 3-dehydro-Δ4-CDCA metabolite as suggested by mass spectrometry (57). 3-dehydro-UDCA did not appear to be a substrate of the baiCD gene product. Bile acid 7α-hydroxy groups are axial while 7β-hydroxy groups are equatorial. These data suggest that the key to substrate recognition between the baiCD and baiH is the stereochemistry of the 7-hydroxy group, whose orientation apparently requires a different architecture in the substrate binding pocket. Taken together these results suggest these homologous enzymes act at an analogous step in the 7-dehydroxylation pathway, and therefore, two distinct branches of the oxidative arm of the pathway exist in C. scindens and C. hiranonis.
The BaiH is a homotrimer composed of 72 kDa subunits flavoprotein containing 1 mol of FMN and 1 mol of FAD per subunit. BaiH is homologous to a class of flavoenzymes involved in reduction of unsaturated fatty acids and aldehydes (Table II) (34). Enoate reductase (enr) from Clostridium tyrobutyricum catalyzes the reduction of C-C double bonds of several α/β-unsaturated aldehydes as well as cyclic ketones and methylketones (52). Enr is a dodecamer with subunit Mr of 73 kDa each containing a 4Fe-4S cluster and 1 mol of FMN and 1 mol of FAD (58). 2, 4-dienoyl-CoA reductase from E. coli is a 73 kDa monomeric enzyme that reduces C-C double bonds with NADPH and contains FMN, FAD as well as a 4Fe-4S cluster (46, 59). The noxB gene from Archaeoglobus fulgidus encodes an NADH oxidase of unknown physiological function (55). The noxB gene product is a 69 kDa monomer containing 1 mol FAD and shares the same conserved ferrodoxin domain (C-2X-C-2-3X-C-11-12X-C) with each of these proteins including the baiH and baiCD gene products (55). The noxB-2 gene shares 98.9 % identity with the noxB-1 gene from A. fulgidus and is found upstream of medium-chain acyl-CoA ligase in this bacterium suggesting these genes are also involved in fatty acid metabolism (55). The more distantly related Old Yellow Enzyme (OYE) family homologues are found both in eukaryotes such as Saccaromyces cerevisiae (OYE1, OYE2) (56) and S. carlsbergensis (OYE1) (60), as well as the prokaryotes including Bacillus subtilis (YqjM) (61), and Shewanella oneidensis (SYE1-SYE4) (62). OYE genes have been extensively characterized both biochemically and structurally (58, 61, 63). Numerous compounds act as electron acceptors including various quinones, as well as α, β-unsaturated aldehydes and ketones which are reduced at the olefinic bond (63); however, only recently have physiologically relevant electron acceptors been identified. The oye2 and oye3 gene from Sac. cerevisiae, sye1 from She. oneidensis, and the yqjM gene from B. subtilis were shown to be inducible by small α/β-unsaturated aldehydes during oxidative stress responses to lipid peroxidation (47, 62, 64) and actin cytoskeletal oxidative damage in yeast (65). Detoxification does not appear to be the function of the bile acid 7α/β-dehydroxylating pathways, but rather the substrates appear to serve as electron sinks to enhance fermentation (2, 14).
Electron flow in the E. coli DCR as determined by redox titrations (66), and crystallography (46) show that two reducing equivalents are supplied to FAD by direct hydride transfer and follow an electron flow from FAD to a 4Fe-4S cluster to FMN and finally to the oxidized substrate. OYEs reduce C=C double bonds through a ping-pong bi-bi mechanism in which FMN is reduced by NADPH, followed by substrate binding and hydride transfer from FMNH2 to the Cβ carbon of the substrate (67). Additional studies will be required to determine the mechanism of the reaction catalyzed by the baiCD and baiH gene products.
We observed low NAD(H)-dependent 3-oxo-Δ4-cholenoic acid oxidoreductase activity based on the amount of products formed, as observed by autoradiography of TLC (Figure 2). However, we would predict that the physiological substrates for these enzymes are the CoA conjugates of 3-oxo-cholanoic acids. This is based on the observations that the baiA gene product encodes a 3α-hydroxysteroid dehydrogenase that has high specificity for coenzyme A conjugated bile acids (28). However, large quantities of the CoA derivatives of 3-dehydro-CDCA and 3-dehydro-UDCA will have to be chemically synthesized to test this hypothesis. In this regard, we have detected NAD(H)-dependent 3-oxo-Δ4-cholenoic acid oxidoreductase activity using small quantities of enzymatically generated radiolabeled CoA conjugates of 3-dehydro-UDCA and 3-dehydro-CDCA (D. Kang, J.M. Ridlon, and P.B. Hylemon, unpublished data). Moreover, other members of this gene family recognize substrates linked to CoA (Table II). Future mechanistic and structural studies will require chemical synthesis of CoA conjugated bile acid intermediates.
This is the first report of a gene product specifically involved in 7β-dehydroxylation of UDCA. We predict that the baiI gene encodes a bile acid 7β-dehydratase, based on its amino acid sequence identity (20 %) and similarity (40 %) with the baiE gene product as well as co-expression of this gene product during CA induction in C. scindens VPI. We have previously demonstrated that the baiE gene encodes a bile acid 7α-dehydratase (33). Determining the function of the baiI as well as identifying genes involved in reducing the 3-oxo-4, 6-choladienoic structure to LCA will be the focus of future of research.
In summary, we have provided data that indicate the baiCD and baiH genes encode NAD(H)-dependent 3-oxo-Δ4-cholenoic acid oxidoreductases. These results can account for the ability of this organism to use UDCA as a substrate for 7β-dehydroxylation, and provide additional support for formation of a 3-oxo-Δ4-cholenoic acid intermediate during bile acid 7-dehydroxylation.
Acknowledgments
This work was supported by an NIH grant PO1-DK38030, R01-DK46390, Shared Instrumentation Grant from NCRR (S10 RR-19231), and Mass Spectrometry Shared Facility grant (P30 CA-13148). We wish to thank Dr. Alan F. Hofmann for his advise during the preparation of this manuscript.
Footnotes
Data presented at Digestive Disease Week 2006, Los Angeles CA (Ref. 57).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vlahcevic ZR, Heuman DM, Hylemon PB. Physiology and Pathophysiology of enterohepatic circulation of bile acids. In: Zakim D, Boyer T, editors. Hepatology: A Textbook of Liver Diseases. 3. Vol. 1. W. B. Saunders; Philadelphia, PA: 1996. pp. 376–417. [Google Scholar]
- 2.Ridlon JM, Kang D, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Bayerdörffer E, Mannes GA, Richter WO, Ochsenkühn T, Wiebecke B, Kopcke W, Paumgartner G. Increased serum deoxycholic acid levels in men with colorectal adenomas. Gastroenterology. 1993;104(1):145–151. doi: 10.1016/0016-5085(93)90846-5. [DOI] [PubMed] [Google Scholar]
- 4.Reddy BS, Wynder EL. Metabolic epidemiology of colon cancer. Fecal bile acids and neutral sterols in colon cancer patients and patients with adenomatous polyps. Cancer. 1977;39:2533–2539. doi: 10.1002/1097-0142(197706)39:6<2533::aid-cncr2820390634>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 5.Hill MJ. Bile flow and colon cancer. Mutat Res. 1990;238:13–32. doi: 10.1016/0165-1110(90)90023-5. [DOI] [PubMed] [Google Scholar]
- 6.Narisawa T, Magadia NE, Weisburger JH, Wynder EL. Promoting effect of bile acids on colon carcinogenesis after intrarectal instillation of N-methyl-N’-nitro-N-nitrosoguanidine in rats. J Natl Cancer Inst. 1974;53(4):1093–1097. doi: 10.1093/jnci/53.4.1093. [DOI] [PubMed] [Google Scholar]
- 7.Reddy BS, Narasawa T, Weisburger JH, Wynder EL. Promoting effect of sodium deoxycholate on colon adenocarcinomas in germfree rats. J Natl Cancer Inst. 1976;56(2):441–442. doi: 10.1093/jnci/56.2.441. [DOI] [PubMed] [Google Scholar]
- 8.Zusman I, Chevion M, Kitrosski N. Effects of N’methyl-N’-nitro-N-nitrosoguanidine and deoxycholic acid on the content of free radicals in rat serum. Exp Toxicol Pathol. 1992;44(4):187–189. doi: 10.1016/S0940-2993(11)80205-8. [DOI] [PubMed] [Google Scholar]
- 9.Cheng K, Raufman J. Bile acid-induced proliferation of a human colon cancer cell line is mediated by transactivation of epidermal growth factor receptors. Bioch Pharm. 2005;70:1035–1047. doi: 10.1016/j.bcp.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 10.Craven PA, Pfanstiel J, DeRubertis FR. Role of activation of protein kinase C in the stimulation of colonic epithelial proliferation and reactive oxygen formation by bile acids. J Clin Invest. 1987;79:532–541. doi: 10.1172/JCI112844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ochsenkuhn T, Bayerdörffer E, Meining A, Schinkel M, Thiede C, Nussler V, Sackmann M, Hatz R, Neubauer A, Paumgartner G. Colonic mucosal proliferation is related to serum deoxycholic acid levels. Cancer. 1999;85(8):1664–1669. [PubMed] [Google Scholar]
- 12.Pai R, Tarnawski AS, Tran T. Deoxycholic acid activates beta-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol Biol Cell. 2004;15(5):2156–2163. doi: 10.1091/mbc.E03-12-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pérez-Ramos P, Olmo N, Turnay J, Lecona E, González de Buitrago G, Portolés MT, Lizarbe MA. Effect of bile acids on butyrate-sensitive and –resistant human colon adenocarcinoma cells. Nutr Cancer. 2005;53(2):208–219. doi: 10.1207/s15327914nc5302_10. [DOI] [PubMed] [Google Scholar]
- 14.Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29(4):625–65. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Maton PN, Murphy GM, Dowling RH. Ursodeoxycholic acid treatment of gallstones. Dose-response study and possible mechanism of action. Lancet. 1977;2:1297–1301. doi: 10.1016/s0140-6736(77)90358-0. [DOI] [PubMed] [Google Scholar]
- 16.Salen G, Colalillo A, Verga D, Bagan E, Tint GS, Shefer S. Effect of high and low doses of ursodeoxycholic acid on gallstone dissolution in humans. Gastroenterology. 1980;78:1412–1418. [PubMed] [Google Scholar]
- 17.Tint GS, Salen G, Colalillo A, Graber D, Verga D, Speck J, Shefer S. Ursodeoxycholic acid: a safe and effective agent for dissolving cholesterol gallstones. Ann Intern Med. 1982;97:351–356. doi: 10.7326/0003-4819-97-3-351. [DOI] [PubMed] [Google Scholar]
- 18.Pares A, Caballeria L, Rodes J. Excellent long-term survival in patients with primary biliary cirrhosis and biochemical response to ursodeoxycholic acid. Gastroenterology. 2006;130(3):715–720. doi: 10.1053/j.gastro.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 19.Earnest DL, Holubec H, Wali RK, Jolley CS, Bissonette M, Bhattacharyya AK, Roy H, Khare S, Brasitus TA. Chemoprevention of azoxymethane-induced colonic carcinogenesis by supplemental dietary ursodeoxycholic acid. Cancer Res. 1994;54(19):5071–5074. [PubMed] [Google Scholar]
- 20.Im E, Martinez JD. Ursodeoxycholic acid (UDCA) can inhibit deoxycholic acid (DCA)-induced apoptosis via modulation of EGFR/Raf-1/ERK signaling in human colon cancer cells. J Nutr. 2004;134:483–486. doi: 10.1093/jn/134.2.483. [DOI] [PubMed] [Google Scholar]
- 21.Khare S, Cerda S, Wali RK, von Lintig FC, Tretiakova M, Joseph L, Stoiber D, Cohen G, Nimmagadda K, Hart J, Sitrin MD, Boss GR, Bissonnette M. Ursodeoxycholic acid inhibits Ras mutations, wild-type Ras activation, and cyclooxygenase-2 expression in colon cancer. Cancer Res. 2003;63(13):3517–23. [PubMed] [Google Scholar]
- 22.White BA, Fricke RJ, Hylemon PB. 7β-Dehydroxylation of ursodeoxycholic acid by whole cells and cell extracts of the intestinal anaerobic bacterium Eubacterium species VPI 12708. J Lipid Res. 1982;23:145–153. [PubMed] [Google Scholar]
- 23.Hirano S, Masuda N. Epimerization of the 7-hydroxyl group of bile acids by the combination of two kinds of microorganisms with 7α- and 7β-hydroxysteroid dehydrogenase activity, respectively. J Lipid Res. 1981;22:1060–1068. [PubMed] [Google Scholar]
- 24.Lepercq P, Gerard P, Beguet F, Raibaud P, Grill JP, Relano P, Cayuela C, Juste C. Epimerization of chenodeoxycholic acid to ursodeoxycholic acid by Clostridium baratii isolated from human feces. FEMS Microbiol Lett. 2004;235(1):65–72. doi: 10.1016/j.femsle.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Sutherland JD, Macdonald IA. The metabolism of primary, 7-oxo- and 7β-hydroxy bile acids by Clostridium absonum. J Lipid Res. 1982;23:726–732. [PubMed] [Google Scholar]
- 26.Mallonee DH, Hylemon PB. Sequencing and expression of a gene encoding a bile acid transporter from Eubacterium sp. strain VPI 12708. J Bacteriol. 1996;178(24):7053–7058. doi: 10.1128/jb.178.24.7053-7058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallonee DH, Adams JL, Hylemon PB. The bile acid-inducible baiB gene from Eubacterium sp. strain VPI 12708 encodes a bile acid enzyme A ligase. J Bacteriol. 1992;174:2065–2071. doi: 10.1128/jb.174.7.2065-2071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallonee DH, White WB, Hylemon PB. Expression in E. coli and characterization of a bile acid-inducible 3α-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. Curr Microbiol. 1995;30:259–263. doi: 10.1007/BF00295498. [DOI] [PubMed] [Google Scholar]
- 29.Engemann C, Elssner T, Pfeifer S, Krumbholz C, Maier T, Kleber H-P. Identification and functional characterization of genes and corresponding enzymes involved in carnitine metabolism of Proteus sp. Arch Microbiol. 2005;183:176–189. doi: 10.1007/s00203-005-0760-2. [DOI] [PubMed] [Google Scholar]
- 30.Heider J. A new family of CoA-transferases. FEBS Letters. 2001;509:345–349. doi: 10.1016/s0014-5793(01)03178-7. [DOI] [PubMed] [Google Scholar]
- 31.Stenmark P, Gurmu D, Nordlund P. Crystal structure of CiaB Type-III CoA transferase in carnitine metabolism. Biochemistry. 2004;43:13996–14003. doi: 10.1021/bi048481c. [DOI] [PubMed] [Google Scholar]
- 32.Ye HQ, Mallonee DH, Wells JE, Björkhem I, Hylemon PB. The bile acid-inducible baiF gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A hydrolase. J Lipid Res. 1999;40:17–23. [PubMed] [Google Scholar]
- 33.Dawson JA, Mallonee DH, Björkhem I, Hylemon PB. Expression and characterization of a C24 bile acid 7α-dehydratase from Eubacterium sp. strain VPI 12708. J Lipid Res. 1996;37:1258–1267. [PubMed] [Google Scholar]
- 34.Baron SF, Hylemon PB. Expression of the bile acid-inducible NADH:flavin oxidoreductase gene of Eubacterium sp. VPI 12708 in Escherichia coli. Biochim Biophys Acta. 1995;1249(2):145–154. doi: 10.1016/0167-4838(95)00034-r. [DOI] [PubMed] [Google Scholar]
- 35.Franklund CV, Baron SF, Hylemon PB. Characterization of baiH gene encoding a bile acid-inducible NADH:Flavin oxidoreductase from Eubacterium sp. strain VPI 12708. J Bacteriol. 1993;175:3002–3012. doi: 10.1128/jb.175.10.3002-3012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White BA, Lipsky RH, Fricke RJ, Hylemon PB. Bile acid induction specificity of 7α-dehydroxylase activity in an intestinal Eubacterium species. Steroids. 1980;35:103–109. doi: 10.1016/0039-128x(80)90115-4. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 39.Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 40.Macdonald IA, Hutchison DM, Forrest TP. Formation of urso- and ursodeoxy-cholic acids from primary bile acids by Clostridium absonum. J Lipid Res. 1981;22:458–466. [PubMed] [Google Scholar]
- 41.Macdonald IA, Roach PD. Bile salt induction of 7α- and 7β-hydroxysteroid dehydrogenases in Clostrdium absonum. Biochim Biophys Act. 1981;665:262–269. doi: 10.1016/0005-2760(81)90011-4. [DOI] [PubMed] [Google Scholar]
- 42.Notredame C, Higgins D, Heringa J. T-Coffee:A novel method for multiple sequence alignments. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 43.Wells JE, Hylemon PB. Identification and characterization of a bile acid 7α-dehydroxylating operon in Clostridium sp. strain TO-931, a highly active 7α-dehydroxylating strain isolated from human feces. Appl Environ Microbiol. 2000;66:1107–1113. doi: 10.1128/aem.66.3.1107-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 45.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 46.Hubbard PA, Liang X, Schulz H, Kim JP. The crystal structure and reaction mechanism of Escherichia coli 2,4-dienoyl-CoA reductase. J Biol Chem. 2003;278(39):37553–37560. doi: 10.1074/jbc.M304642200. [DOI] [PubMed] [Google Scholar]
- 47.Fitzpatrick TB, Auweter S, Kitzing K, Clausen T, Amrhein N, Macheroux Structural and functional impairment of an Old Yellow Enzyme homologue upon affinity tag incorporation. Protein Expr Purif. 2004;36:280–291. doi: 10.1016/j.pep.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Chen J, Liu J, Yang X, Wang K. Binding of Cu2+ to S-adenosyl-L-homocysteine hydrolase. J Inorg Biochem. 2004;98:977–983. doi: 10.1016/j.jinorgbio.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 49.Sulkowski E. Purification of proteins by IMAC. Trends Biotechnol. 1985;3:1–7. [Google Scholar]
- 50.Hylemon PB, Melone PD, Franklund CV, Lund E, Björkhem I. Mechanism of intestinal 7α-dehydroxylation of cholic acid: evidence that allo-deoxycholic acid is an inducible side-product. J Lipid Res. 1991;32:89–95. [PubMed] [Google Scholar]
- 51.Liu XL, Scopes RK. Cloning, sequencing and expression of the gene encoding NADH oxidase from the extreme anaerobic thermophile Thermoanaerobium brockii. Biochim Biophys Acta. 1993;1174(2):187–190. doi: 10.1016/0167-4781(93)90113-r. [DOI] [PubMed] [Google Scholar]
- 52.Rohdich F, Wiese A, Feicht R, Simon H, Bacher A. Enoate reductases of Clostridia. Cloning, sequencing, and expression. J Biol Chem. 2001;276(8):5779–5787. doi: 10.1074/jbc.M008656200. [DOI] [PubMed] [Google Scholar]
- 53.Bao Q, Tian Y, Li W, Xu Z, Xuan Z, Hu S, Dong W, Yang J, Chen Y, Xue Y, Xu Y, Lai X, Huang L, Dong X, Ma Y, Ling L, Tan H, Chen R, Wang J, Xu J, Yang H. A complete sequence of the T. tengcongensis genome. Genome Res. 2002;12(5):689–700. doi: 10.1101/gr.219302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Björkhem I, Einarsson K, Melone PD, Hylemon PB. Mechanism of intestinal formation of deoxycholic acid from cholic acid in humans: evidence for a 3-oxo-Δ4-steroid intermediate. J Lipid Res. 1989;30:1033–1039. [PubMed] [Google Scholar]
- 55.Kengen SW, van der Oost J, de Vos WM. Molecular characterization of H2O2-forming NADH oxidases from Archaeoglobus fulgidus. Eur J Biochem. 2003;270:2885–2894. doi: 10.1046/j.1432-1033.2003.03668.x. [DOI] [PubMed] [Google Scholar]
- 56.Stott K, Saito K, Thiele DJ, Massey V. Old yellow enzyme: the discovery of multiple isozymes and a family of related proteins. J Biol Chem. 1993;268(9):6097–6106. [PubMed] [Google Scholar]
- 57.Ridlon JM, Hylemon PB, Kang D, Mallonee D, Wells JE, Moore DR, Barnes S. The baiCD and baiH genes from Clostridium scindens encode 3-oxo-Δ4-steroid oxidoreductases specific for 7α and 7β-hydroxy bile acids, respectively. Gastroenterology. 2006;130(4 Suppl 2):A–829. [Google Scholar]
- 58.Kuno S, Bacher A, Simon H. Structure of enoate reductase from a Clostridium tyrobutyricum (C. spec. La1) Biol Chem Hoppe-Seyler. 1985;366:463–472. doi: 10.1515/bchm3.1985.366.1.463. [DOI] [PubMed] [Google Scholar]
- 59.Dommes V, Kunau WH. 2,4-Dienoyl coenzyme A reductases from bovine liver and Escherichia coli. Comparison of properties. J Biol Chem. 1984;259:1781–1788. [PubMed] [Google Scholar]
- 60.Saito K, Thiele DJ, Davio M, Lockridge O, Massey V. The cloning and expression of a gene encoding Old Yellow Enzyme from Saccharomyces carlsbergensis. J Biol Chem. 1991;266(31):20720–20724. [PubMed] [Google Scholar]
- 61.Kitzing K, Fitzpatrick TB, Wilken C, Sawa J, Bounekov GP, Macheroux P, Clausen T. The 1.3 Å crystal structure of the flavoprotein YqjM reveals a novel class of Old Yellow Enzymes. J Biol Chem. 2005;280(30):27904–27913. doi: 10.1074/jbc.M502587200. [DOI] [PubMed] [Google Scholar]
- 62.Brigé A, van den Hemel D, Carpentier W, De Smet L, Van Beeumen JJ. Comparative characterization and expression analysis of the four Old Yellow Enzyme homologues from Shewanella oneidensis indicate differences in physiological function. Biochem J. 2006;394:335–344. doi: 10.1042/BJ20050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaz ADN, Chakraborty S, Massey V. Old yellow enzyme: aromatization of cyclic enones and the mechanism of a novel dismutation reaction. Biochemistry. 1995;34:4246–4256. doi: 10.1021/bi00013a014. [DOI] [PubMed] [Google Scholar]
- 64.Trotter EW, Collinson EJ, Dawes IW, Grant CM. Old yellow enzymes protect against acrolein toxicity in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol. 2006;72(7):4885–4892. doi: 10.1128/AEM.00526-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haarer BK, Amberg DC. Old yellow enzyme protects the actin cytoskeleton from oxidative stress. Mol Biol Cell. 2004;15:4522–4531. doi: 10.1091/mbc.E04-06-0445. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Liang X, Thorpe C, Schulz H. 2,4-Dienoyl-CoA reductase from Escherichia coli is a novel iron-sulfur flavoprotein that functions in fatty acid beta-oxidation. Arch Biochem Biophys. 2000;380(2):373–379. doi: 10.1006/abbi.2000.1941. [DOI] [PubMed] [Google Scholar]
- 67.Brown BJ, Deng Z, Karplus PA, Massey V. On the active site of Old Yellow Enzyme. Role of histidine 191 and asparagine 194. J Biol Chem. 1998;273(49):32753–32762. doi: 10.1074/jbc.273.49.32753. [DOI] [PubMed] [Google Scholar]



