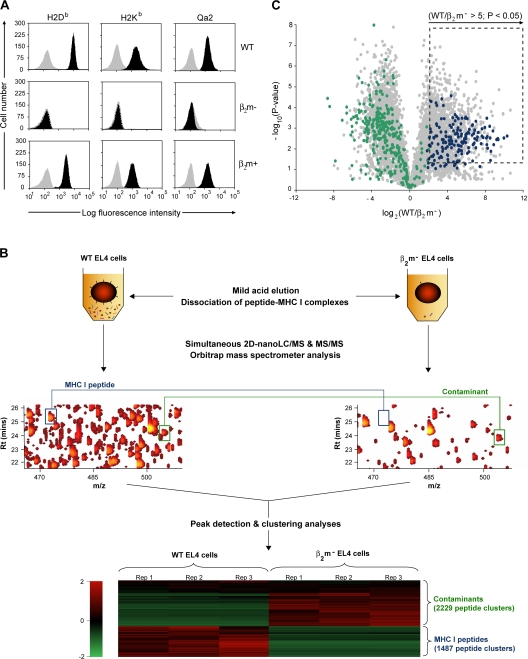
Figure 1.
Experimental design for identification and relative quantification of native unlabeled MHC I–associated peptides. (A) Cell surface MHC I expression on EL4 WT, β2m− mutant, and β2m+ transfectant cell lines. Cells were stained with antibodies against H2Db, H2Kb, and Qa2 (black) or the corresponding isotype control antibody (gray). (B) Peptides obtained by MAE were analyzed by on-line 2D-nanoLC-MS (three biological replicates for each cell population). Contour profiles of m/z versus retention time versus intensity were used to visualize differences between MS profiles (middle). A logarithmic intensity scale distinguishes between low (dark red) and highly (bright yellow) abundant species. Examples of peptides that were differentially expressed (blue line) or not (green line) between WT and β2m− EL4 cells are highlighted in boxes. (bottom) Heat map representation shows differential peptide expression between WT and β2m− EL4 cells where each horizontal line corresponds to a unique peptide cluster (n = 3,716). A logarithmic scale depicts peptides that are expressed at high (red) or low (green) level in each cell population. (C) Volcano plot representation showing reproducibly detected peptide ions across three replicate analyses. Peptide clusters (n = 1,236) highlighted in dashed box were considered as class I–restricted (P ≤ 0.05; fold change ≥ 5). Peptides that were sequenced by MS/MS are represented by colored dots: green for contaminants, and blue for MHC I–associated peptides.