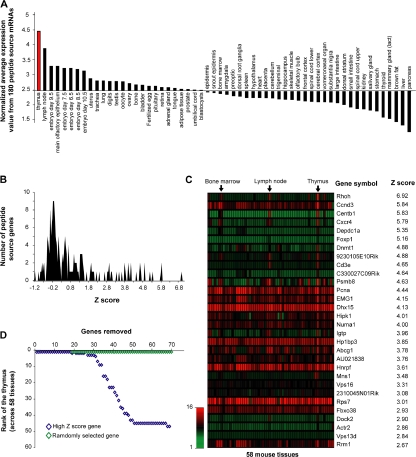
Figure 5.
Peptide source mRNAs expression patterns reveal an organ-specific signature in the MIP repertoire of thymocytes. (A) Comparison of normalized mean expression values (y axis) across 58 different tissues (x axis) including the thymus (red). Normalized mean expression values were calculated as follows: mean expression value from 180 peptide source genes/mean expression value of 36,182 transcripts for each particular tissue. Calculated values were ranked from left to right in a decreasing order. (B) Z scores were calculated for each of the 180 peptide source genes to identify transcripts preferentially expressed in the thymus. Graph shows frequency distributions of calculated z scores with a bin increment of 0.05. (C) Heat map shows the relative mRNA expression in 58 tissues of the 30 peptide source genes with highest thymic z scores. (D) High z score genes determine the thymus specificity of the gene set encoding MHC I–associated peptides. Genes preferentially overexpressed in the thymus (high thymic z scores; blue) were additively removed (x axis). Normalized mean expression values and thymus rank (y axis) were determined following removal of each individual gene. Removal of the 30 genes with a high thymic z score had a drastic impact on thymus rank. Removal of up to 70 randomly selected genes (100,000 permutations; green) had no significant impact on thymus rank.