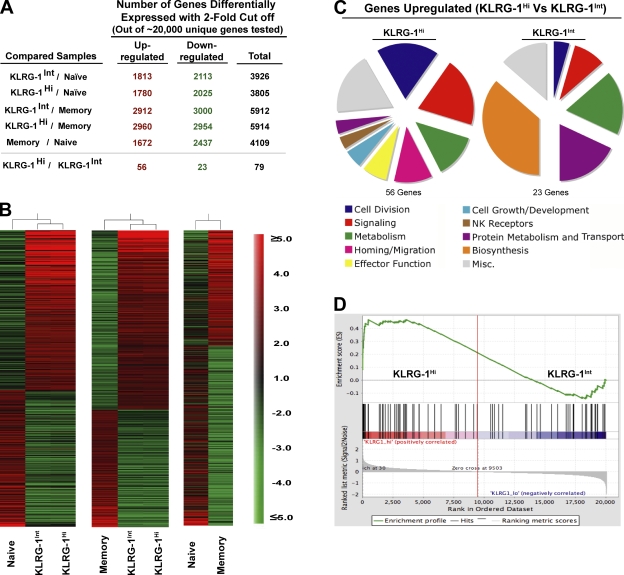
Figure 5.
Genome-wide microarray analysis of memory precursors and terminal effectors. Expression levels of mRNA were measured by microarray analysis of sorted P14 CD44Lo naive cells, KLRG-1Int and KLRG-1Hi effector cells at day 4.5 p.i., and P14 memory cells at day 60 p.i. Gene ratios (fold change) represent the mean value of three to four independent hybridizations to Affymetrix microarrays. (A) Total number of transcripts differentially expressed (up- or down-regulated; twofold cutoff) between the indicated comparison groups is shown. (B) Relative intensities of all genes with a 2.5-fold cutoff in KLRG-1Int and KLRG-1Hi cells with respect to naive and memory populations are plotted as heat maps to depict the relationship between various populations. (C) All genes differentially expressed (twofold cutoff) between KLRG-1Int and KLRG-1Hi cells are functionally classified under broad categories based on information found in the Gene Ontology and Ingenuity Pathways databases and plotted as pie charts. (D) Enrichment profile of effector signature genes in KLRG-1Hi versus KLRG-1Int effector cells. GSEA of a set of genes corresponding to differentiated effectors (D8) in the rank-ordered list of genes differentially expressed between KLRG-1Int versus KLRG-1Hi effector cells is shown.