Figure 4. .
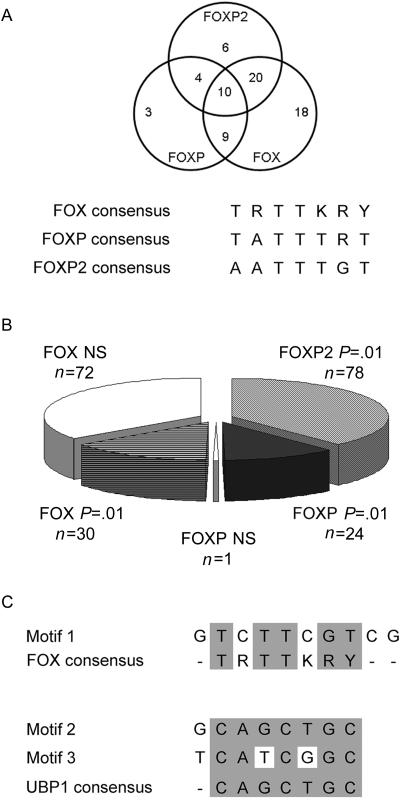
In silico motif analysis of sequences enriched in FOXP2 ChIP-chip. A, Assessment of the presence of known FOX (TRTTKRY), FOXP (TRTTTRY), or FOXP2 ([A]ATTTG[T]) binding sites in the promoter regions of the genes in table 3. The Venn diagram represents the number of promoter sequences containing at least one of these binding sites, including overlaps where more than one class of site is present in the same sequence. Of the promoters, 70 contained at least one perfect FOXP2, FOXP, or FOX binding site, with most (61%) containing multiple classes of consensus sites. All promoters featured at least one FOXP2 binding site that contained a single mismatch. B, Number and significance of binding sites. The total number of binding sites identified in all sequences is represented, partitioned according to the site class and the statistical significance of identifying each site within the data set. NS = not significant. C, Results of unbiased motif analysis. Overrepresented motifs identified from the 100 target sequences analyzed included motif 1, which closely resembled the FOX family consensus binding site, and motifs 2 and 3, which were close or exact matches to the UBP1 (also known as “LBP-1a”) consensus binding site.