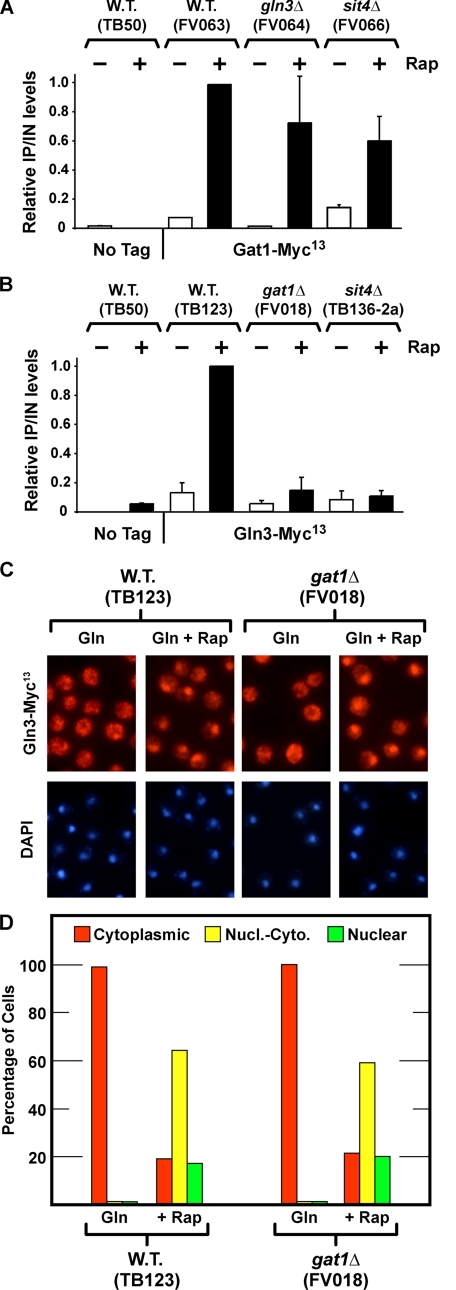
FIGURE 4.
A and B, effects of sit4Δ, gln3Δ, and gat1Δ on rapamycin-induced binding of Gat1-Myc13 and Gln3-Myc13 to the DAL5 promoter. C and D, effect of deleting GAT1 on Gln3-Myc13 localization. Wild type (W.T.) untagged (TB50), wild type GAT1-MYC13 (FV063), gln3Δ GAT1-MYC13 (FV064), and sit4Δ GAT1-MYC13 (FV066) (A), and wild type untagged (TB50), wild type GLN3-MYC13 (TB123), gat1Δ GLN3-MYC13 (FV018), sit4Δ GLN3-MYC13 (TB136-2a) (B) strains were grown in YNB-glutamine medium with or without the addition of rapamycin (0.2 μg/ml) for 30 min. ChIP was performed using antibodies against c-Myc as described under “Materials and Methods.” Quantitative PCR of IP and IN fractions was performed with primers for DAL5 promoter (DAL5P) and for a region 2.5 kb upstream of DAL5 open reading frame as a control (DAL5U). For each immunoprecipitation, IP/IN values were calculated as follows: [DAL5P]IP/[DAL5P]IN – [DAL5U]IP/[DAL5U]IN, normalized to the value obtained with wild type-induced cells. Histograms represent the average of at least two experiments from independent cultures. The error bars indicate standard errors. C and D, wild type and gat1Δ strains were grown in YNB-glutamine medium. Split cultures were left untreated (Gln) or treated with rapamycin (0.2 μg/ml) for 20 min (+Rap), sampled, and processed for immunofluorescence microscopy as described under “Materials and Methods” and in the legend to Fig. 3.