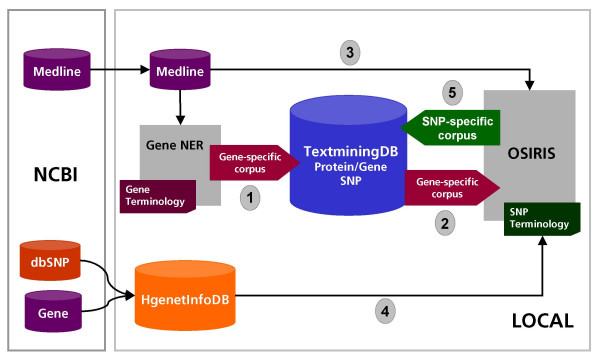
Figure 1.
OSIRISv1.2 system work flow. The system is based on a local mirror of MEDLINE database and HgenetInfoDB, which integrates information for human Gene and dbSNP databases from the NCBI. The starting point of the system is a gene, for which a set of articles is annotated using the NER tool ProMiner and stored in the TextMiningDB (1). The gene-specific corpus is accessed by OSIRISv1.2 (2) to obtain the MEDLINE citations annotated to a NCBI Gene entry. The corresponding MEDLINE abstracts are retrieved from a local repository (3). In addition, sequence data for each gene and its sequence variants are retrieved from HgenetInfoDB, and this information is used to generate the SNP terminology (4). The next step of OSIRISv1.2 is the search for occurrences of the sequence variant terms in each gene-specific corpus by processing the MEDLINE abstracts. This information (SNP-specific corpus) is returned to the TextMiningDB database (5). The results of OSIRISv1.2, stored in the TextMiningDB, can be accessed through our web interface at [25]. GenDB: data retrieval system used for conversion of the XML files to the files in MEDLINE format, indexing of the MEDLINE files and for their retrieval (internal development of FhG-SCAI by Theo-Heinz Mevissen). ProMiner has been described elsewhere [15]. For simplicity we use in this figure the term SNP to refer to all variations present in the database (SNPs and other types of sequence variants).