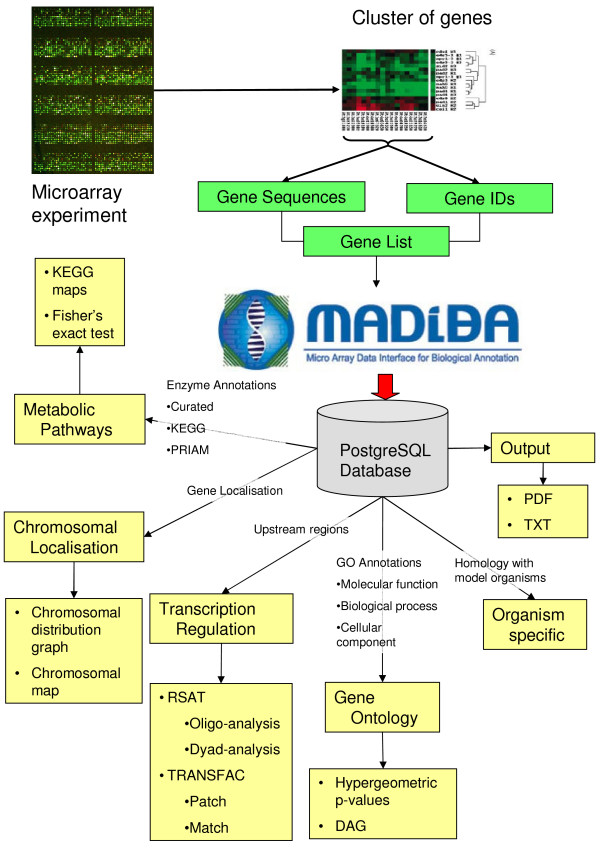
Figure 1.
A schematic representation of the flow of data through MADIBA. After a microarray experiment, data are normalised and then clustered, since it is hypothesised that the genes in a cluster have common biological implications. A cluster of genes is submitted to MADIBA, either as nucleotide sequences, or gene identifiers. This list of genes can then be subjected to five analysis modules – Gene Ontology Analysis, Metabolic Pathways Analysis, Transcription Regulation Analysis, Chromosomal Localisation Analysis and an Organism Specific Analysis. Also shown are the data that are required by each of the analysis modules. The results from the analyses can be exported as a PDF file, or as plain text.