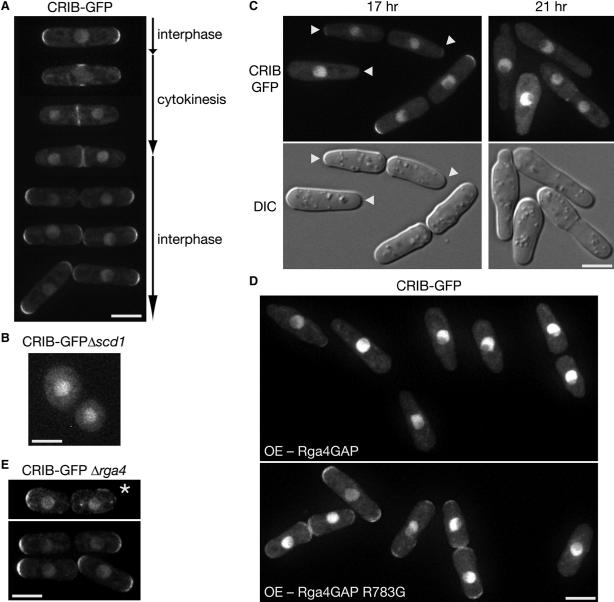
Figure 4. Rga4 Is Involved in Localization of Active Cdc42.
(A) Fluorescence microscopy of a wild-type strain expressing CRIB-GFP. Images of cells at different stages of the cell cycle are shown. (B) Fluorescence microscopy of a Δscd1 strain expressing CRIB-GFP. Cortical CRIB-GFP was hardly detectable. (C) Fluorescence (CRIB-GFP; top) and DIC (bottom) microscopy of a strain overexpressing Rga4. A wild-type strain expressing CRIB-GFP was transformed with the pREP1-rga4:FLAG plasmid and grown at 30°C for 17 hr and 21 hr in EMM without thiamine to induce Rga4FLAG expression. Arrowheads indicate the growing cell tip with reduced diameter. (D) Wild-type (Rga4GAP) or arginine-finger mutant (Rga4GAP R783G) GAP domains tagged with the FLAG epitope were overexpressed using the pREP1 vector in a CRIB-GFP strain. Cells were cultured in EMM medium without thiamine for 14 hr at 30°C to induce expression from the nmt1 promoter. Anti-FLAG immunoblotting confirmed that the wild-type and mutant Rga4 GAP domain fragments were expressed at similar levels (data not shown). (E) Fluorescence microscopy of a Δrga4 strain expressing CRIB-GFP. An asterisk marks a Δrga4 cell in which CRIB-GFP appears depolarized. Scale bars represent 5 μm. Images were taken at 0.4 μm steps and deconvolved for projection images.