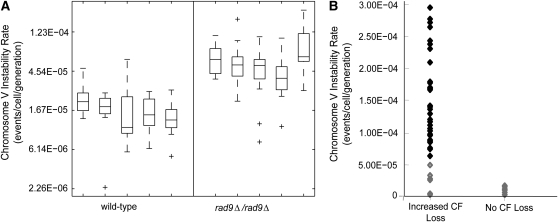
Figure 2.—
Validation of chromosome V instability rate estimates and enrichment of increased CF loss strains for increases in chromosome V instability. (A) Fluctuation analysis experiments were performed on five wild-type and five rad9-deficient yeast strains to test reproducibility of the chromosome V instability rate estimations. These data are shown in a combined box and whisker plot where each box represents summary statistics of instability rate estimates in the 15 parallel cultures of each strain. The box has lines at the lower quartile, median, and upper quartile of the data. The whiskers are lines extending from each end of the box to show the extent of the rest of the data. The whiskers indicate the minimum and maximum data values, unless outliers (marked by “+”) are present in which case the whiskers extend to a maximum of 1.5 times the interquartile range. (B) We measured the chromsome V instability rates of 40 mutant strains from the increased CF loss category and 10 mutant strains from the nonsectoring category. One-sided unequal variance t-tests were performed on the estimated rate of chromosome V instability of each of the 50 strains compared to the rad9Δ/rad9Δ parental strain (3.37E-05 by summation of five separate rad9Δ/rad9Δ estimations). Strains that were statistically increased as determined by the t-test are shown as solid black diamonds; strains not significantly increased are shown as open gray diamonds. Seventy percent of the strains from the high CF loss group showed statistically increased chromosome V instability rates, while none of the strains from the no CF loss group were statistically different.