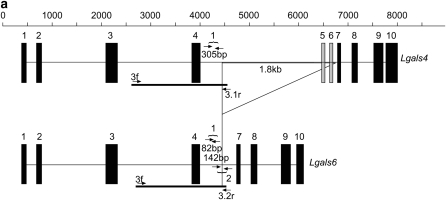
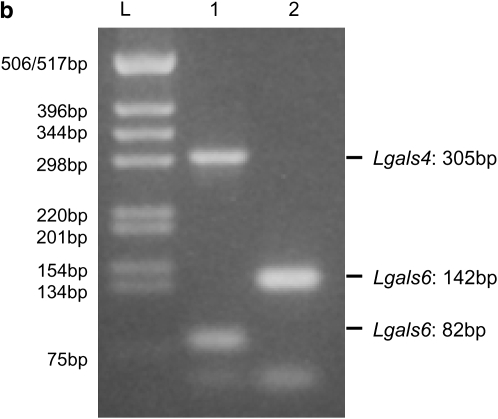
Figure 1.—
Comparison of Lgals4 and Lgals6 genomic organization. (a) Genomic organization of the Lgals4 (top) and Lgals6 (bottom) genes. Exons are represented as boxes, numbered from 01 to 10. Note that, for clarity, we ascribe the same reference number to homologous exons in Lgals4 and Lgals6, i.e., the exons of both genes are numbered from 01 to 10 with exons 05 and 06 (shaded on Lgals4) missing from the Lgals6 gene. Scale bar is in base pairs. The Lgals4 and Lgals6 genes differ by a 1.8-kb deletion in Lgals6 shown here as an open triangle. The primer pair 1f-1r (numbered 1) amplifies a 305-bp fragment specific for the Lgals4 gene and an 82-bp fragment specific for the Lgals6 gene. The primer pair 2f-2r (numbered 2) amplifies a 142-bp fragment specific for the Lgals6 gene. The fragment containing the intronic sequences that were cloned and sequenced to build the phylogenetic tree is shown as a solid line (numbered 3.1 in Lgals4 and 3.2 in Lgals6, respectively). (b) Ethidium bromide-stained gel showing bands amplified from the primer pair 1f-1r (numbered 1) and 2f-2r (numbered 2) from 129sv genomic DNA. L, DNA ladder.