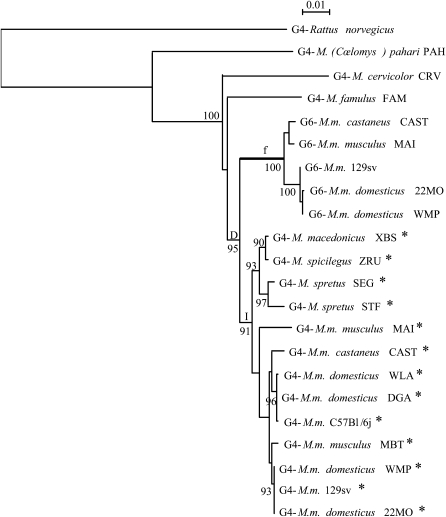
Figure 4.—
Phylogenetic tree of Lgals4 and Lgals6. This maximum-likelihood tree was reconstructed with 1234 sites of intronic sequences (input tree generated by BIONJ; HKY model including a Γ-correction with four categories of sites and ts:tv ratio estimated from the data). The numbers at nodes correspond to the percentage support of 1000 bootstrap replicates and percentages only >90% are shown. The intron 04 of sequences with an asterisk contains the SINE element. The branch f is postulated to be under positive selection and is considered as the foreground branch for branch-site models. D represents the gene duplication event; I, the insertion of the SINE element in the intron 04 of some Mus Lgals4 sequences.