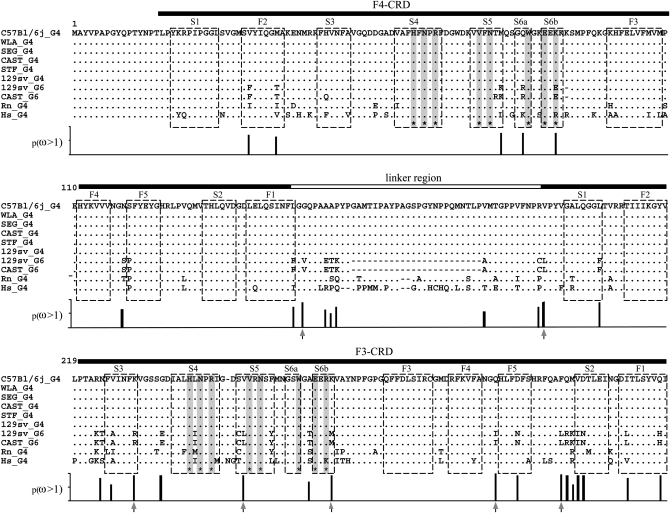
Figure 5.—
Comparison of amino acid sequences of galectin-4 and galectin-6 sequences. Dashes represent gaps introduced for alignment: a genomic deletion in the Lgals6 gene removed two exons (05 and 06; see Figure 1), coding for part of the linker. Residues identical to those of the corresponding C57BL/6J galectin-4 are indicated by dots. Horizontal filled bars correspond to the F4- and F3-carbohydrate recognition domains (Houzelstein et al. 2004). The horizontal open bar corresponds to the linker region that is shorter in galectin-6 (G6) because of the deletion. Asterisks at the bottom of the sequences and corresponding shaded vertical bars mark the position of residues that interact with carbohydrates (Lobsanov et al. 1993). Open boxes indicate the β-strands. Vertical bars below the alignment show the Bayesian posterior probability of ω > 1 for each site. Arrows under vertical bars indicate sites with p(ω >1) > 0.95.