TABLE 2.
Posterior estimates for various levels of genetic differentiation and low gene flow
|
FST
|
|||||
|---|---|---|---|---|---|
| Factors included | 0.01 | 0.05 | 0.10 | 0.25 | |
| None | 0.444 | 0.106 | 0.056 | 0.028 | |
| G1 | 0.122 | 0.072 | 0.055 | 0.030 | |
| G2 | 0.291 | 0.301 | 0.122 | 0.073 | |
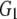 and and 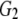
|
0.075 | 0.323 | 0.521 | 0.638 | |
| With interaction | 0.068 | 0.197 | 0.246 | 0.231 | |
| Estimate/RMSE/95% HPDI
|
|||||
| Parameter | True value | 0.01 | 0.05 | 0.10 | 0.25 |
| α1 | −0.900 | a | −0.858 | −1.122 | −1.010 |
| 0.123 | 0.101 | 0.015 | |||
| (−1.984; 0.263) | (−2.091; −0.191) | (−1.723; −0.321) | |||
| α2 | 1.100 | a | 1.403 | 1.314 | 1.173 |
| 0.127 | 0.071 | 0.008 | |||
| (0.197; 2.687) | (0.387; 2.279) | (0.473; 1.886) | |||
| σ2 | – | 0.486 | 0.513 | 0.452 | 0.352 |
| — | — | — | — | ||
| (0.131; 3.067) | (0.125; 4.117) | (0.120; 3.090) | (0.110; 1.956) | ||
| Assignments
|
0.01
|
0.05
|
0.10
|
0.25
|
|
| Misassignments | 0.808 | 0.134 | 0.046 | 0.002 | |
| Probabilitiesb | 0.288 | 0.847 | 0.946 | 0.997 | |
Posterior model probabilities, regression parameter mean estimates, and assignment accuracy for synthetic data generated with proportions of nonmigrant alleles set to mii = 0.90 are shown.
The corresponding regression parameter is not included in the model with the highest posterior model probability.
Maximum posterior assignment probabilities averaged across all individuals.
