TABLE 6.
Posterior model estimates when testing for nonexplanatory factors
|
FST
|
||||
|---|---|---|---|---|
| Factors included | 0.01 | 0.05 | 0.10 | 0.25 |
| None | 0.636 | 0.533 | 0.512 | 0.505 |
| G1 | 0.127 | 0.224 | 0.243 | 0.241 |
| G2 | 0.184 | 0.157 | 0.151 | 0.158 |
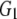 and and 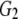
|
0.039 | 0.065 | 0.071 | 0.076 |
| With interaction | 0.013 | 0.022 | 0.022 | 0.021 |
|
FST
|
||||
| Assignments | 0.01 | 0.05 | 0.10 | 0.25 |
| Misassignments | 0.741 | 0.282 | 0.110 | 0.002 |
| Probabilities | 0.218 | 0.694 | 0.881 | 0.996 |
Posterior model probabilities and assignment accuracy when varying the level of genetic differentiation FST and testing for two nonexplanatory factors (i.e., different from the ones we used for generating migration rates) are shown. Data were simulated with proportions of nonmigrant alleles set to mii = 0.70.
