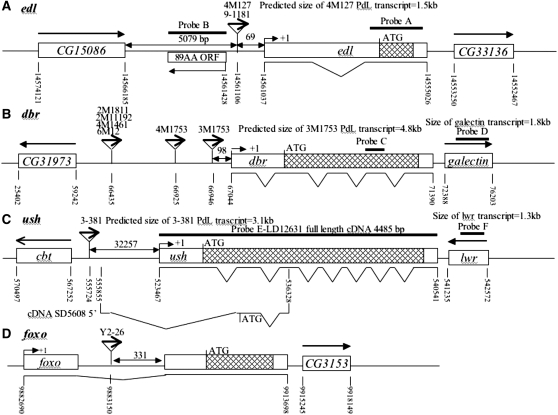
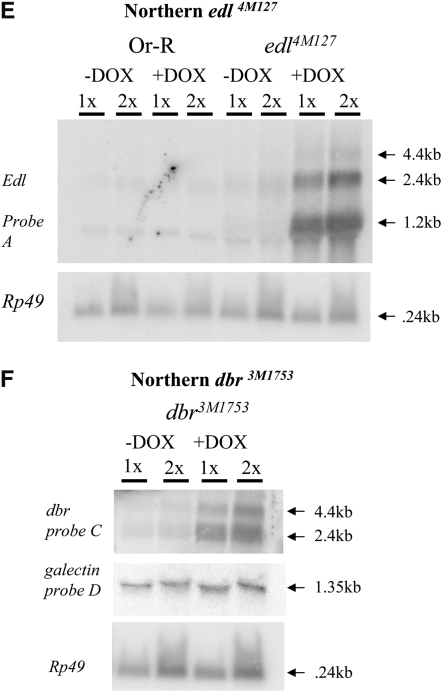
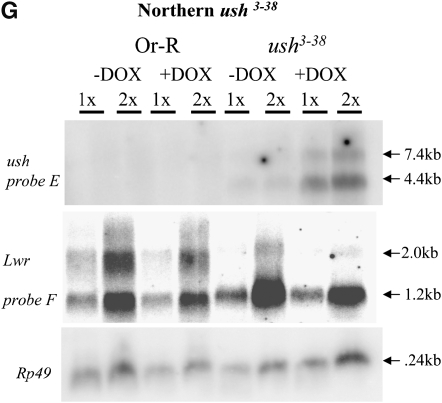
Figure 4.—
Molecular characterization of selected mutations. (A–D) Intron and exon boundaries of the mutated genes are indicated, with numbering according to DNA sequences obtained from the NCBI web site (http://www.ncbi.nlm.nih.gov/). Locations for transcriptional initiation are indicated by arrows. PdL inserts are indicated by triangles and each is oriented 5′ to 3′, as indicated by internal arrows. DNA fragments used as probes in Northern analyses are indicated at the top of each diagram. (A) edl (genomic scaffold sequence AE003798.3). (B) dbr (genomic scaffold sequence AE003590.3). (C) ush (genomic scaffold sequence AE003589.3; SD5608 cDNA sequence is from accession no. AI541608 in the NCBI database. (D) foxo. (E–G) Northern analysis of selected lines. Oregon R wild-type strain and the indicated PdL mutant strains were crossed to rtTA(3)E2 driver strain. Total RNA was isolated from adult progeny cultured 1 week in the presence and absence of DOX, transferred to Northerns blots, and hybridized with the gene-specific probes indicated. Ribosomal protein 49 gene Rp49 was used as control for loading. Two amounts of RNA were loaded for each sample (once and twice, as indicated), and signals were visualized by phosphoimager. (E) edl4M127 mutant strains and controls. (F) dbr3M1753 mutant strain and controls. (G) ush3-38 mutant strains and controls. Galectin probe D downstream of gene dbr and lwr probe F downstream of gene ush did not yield any altered signal in the presence and absence of DOX, indicating that transcription did not go beyond genes dbr and ush. The predicted sizes of the edl4M127, dbr3M1753, and ush3-38 PdL transcripts (A–C) matched the measured sizes of the corresponding +DOX transcripts (E–G).