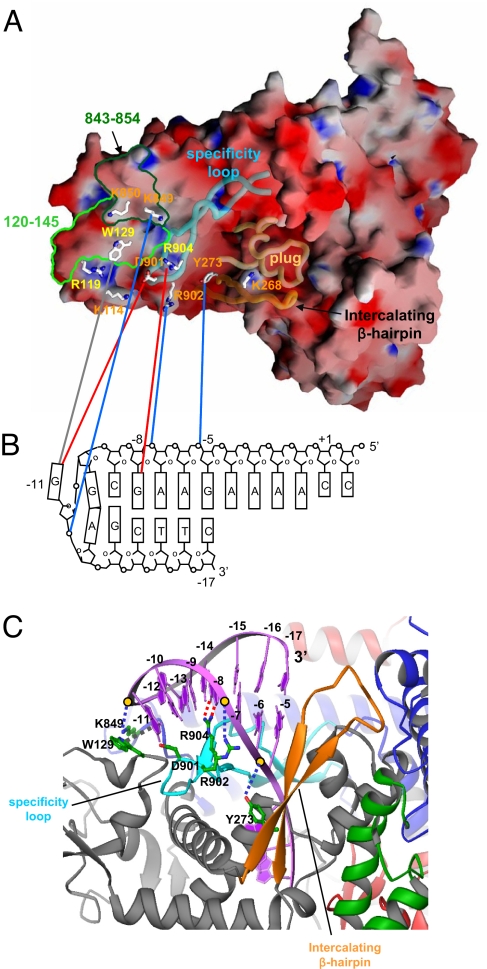
Fig. 4.
Recognition of hairpin-promoter by N4 mini-vRNAP. (A) Electrostatic distribution of N4 mini-vRNAP. Positive electrostatic potential is blue, and negative potential is red. The transparent α-carbon backbones of the specificity loop, intercalating β-hairpin and plug are superimposed. Residues identified from biochemical studies to be important for promoter binding are depicted as stick model and labeled and color-coded as follows: major promoter binding and salt resistance, yellow; minor promoter binding, orange. Mapped −11 5IdU (light green, residues 120–145) and −11/−12 AzPh (dark green, residues 843–854) cross-links are also shown. This figure was made with the program GRASP (32). (B) N4 mini-vRNAP and promoter hairpin DNA interaction. A schematic representation of the N4 vRNAP P2 promoter is shown. The transcription start site (+1), beginning, and end of the hairpin-stem (−5 and −17, respectively), and DNA bases essential for vRNAP binding (−8 and −11) are indicated. N4 mini-vRNAP residues that participate in promoter recognition (A) are connected to their proposed interacting partners on promoter DNA (B) by lines: potential van der Waal's interaction between −11 G and W129 (gray), −8 G recognition by R904 (red), −11 G recognition by D901 (red), and phosphate backbone interactions by Y273 (−5/−6 P), R902 (−7/−8 P) and K849 (−11/−12 P) (blue). (C) Docked model of the N4 mini-vRNAP and promoter hairpin DNA. Hairpin-form DNA is depicted as purple ribbon and bases are labeled. N4 mini-vRNAP residues that participate in promoter recognition are indicated. Dashed lines indicate protein-DNA interactions as shown in B. This figure was made with the program Ribbons (33).