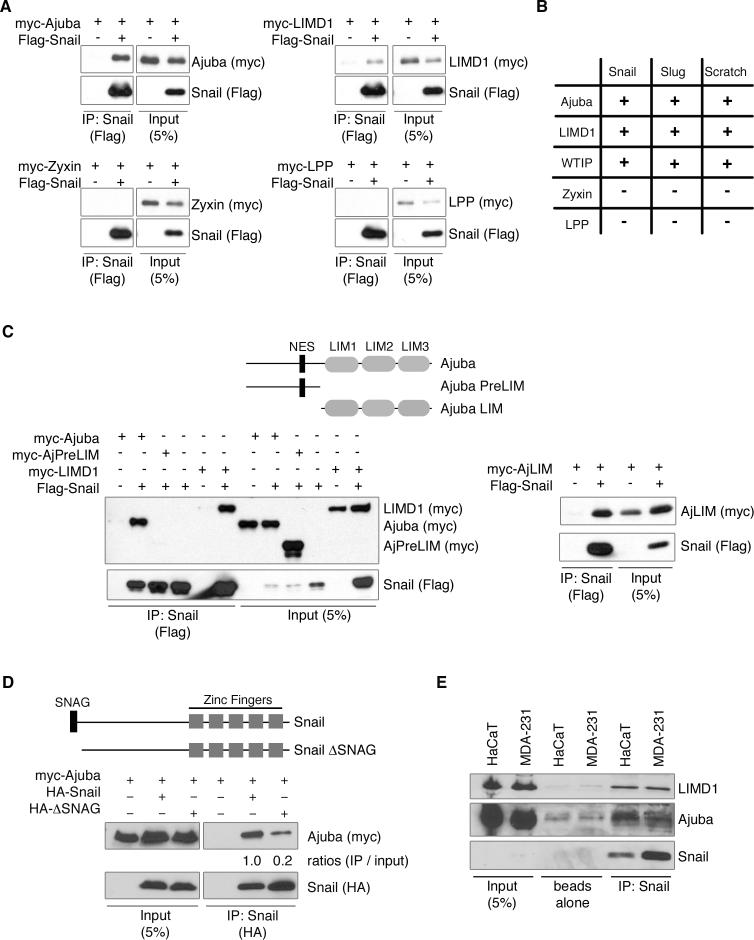
Figure 1. Ajuba LIM proteins interact with Snail transcriptional repressors.
A. Myc-tagged LIM proteins and Flag-Snail were cotransfected into HEK293 cells. Snail was immunoprecipitated (anti-Flag) and bound products Western blotted for LIM protein (anti-myc) and Snail (anti-Flag). Control Western blot of lysate is on right panel of each set. B. Table of interactions between Snail proteins and LIM proteins, as determined by co-immunoprecipitation, as described in A. C. Top: Schematic of Ajuba constructs used. NES – nuclear export sequence. Bottom: Co-immunoprecipitation experiments as in A. D. Top: Schematic of Snail constructs used. Bottom: Myc-tagged Ajuba and HA-tagged Snail constructs were cotransfected into HEK293 cells. Snail was immunoprecipitated (anti-HA) and bound products Western blotted for Ajuba (anti-myc) and Snail (anti-HA). Control Western blot of lysate is shown on left. The amount of Ajuba immunoprecipitated relative to input was quantified by densitometry and controlled for the amount of Snail immunoprecipitated. (Ajuba immunoprecipitated with full-length Snail was arbitrarily set to 1). E. Endogenous Snail was immunoprecipitated from lysates of HaCaT or MDA-231 cells and bound products Western blotted for the presence of Ajuba, LIMD1 and Snail. Controls include pulldown with Protein G beads alone and lysate input.