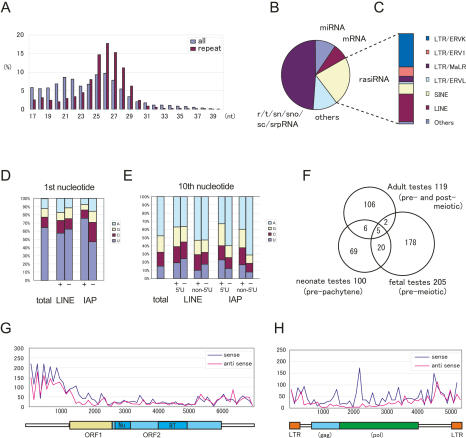
Figure 3.
piRNAs in fetal premeiotic germ cells. (A) In total, 127,997 small RNAs were sequenced from E12.5–E19.5 fetal germ cells. The size distributions (in nucleotides) of the total small RNAs and rasiRNAs are shown by blue and purple bars, respectively. (B,C) Genomic annotation of the small RNAs (B) and the ratio of piRNA sequences of fetal premeiotic germ cells (C). Detailed results are listed in Supplemental Table S2. (D,E) Comparison of the first (D) and tenth (E) nucleotides of the total, sense (+), and antisense (−) piRNAs. Nucleotide biases were calculated for the Gf type Line-1 and IAP piRNAs analyzed in Supplemental Table S2. (E) The piRNA classes that contain or lack a 5′-end U are shown separately. (F) Venn diagram of the piRNAs in adult pre- and post-meiotic (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006), neonatal prepachytene (Aravin et al. 2007), and fetal (this study) testes. (G,H) Distribution of piRNAs corresponding to type Gf Line-1 (G) and IAPIΔ1 (H).