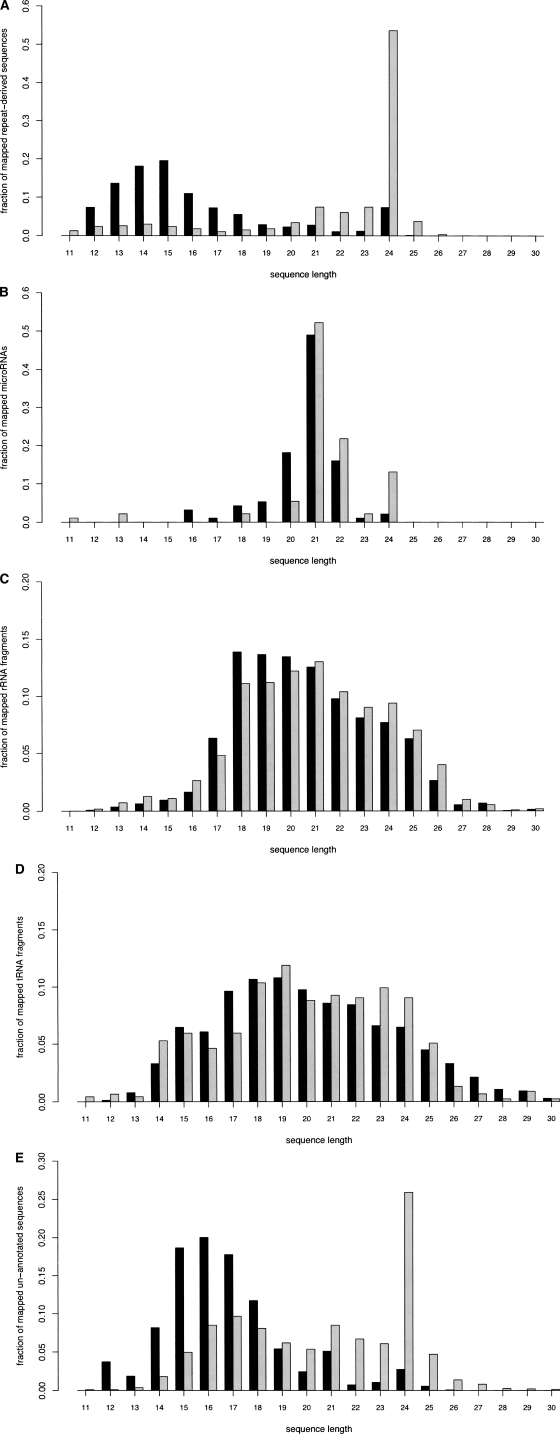
Figure 2.
Length distribution of unique P. contorta (black bars) and O. sativa (gray bars) small RNAs sorted by class and that map perfectly to at least one genomic locus in the O. sativa genome. A total of 5129 unique P. contorta sequences mapped to the O. sativa genome. The Y-axis reflects the fraction of sequences of the stated annotation residing within each length bin. (A) Genomic repeats as annotated by RepeatMasker using RepBase, v. 9.04. Two distinct lobes are observable, the first due mainly to the high probability of shorter sequences occurring by chance in the genome. The second lobe peaks for both species with 24-nt sequences, suggesting an over-representation of conserved sequences that are 24 nt in length. (B) Conserved miRNAs matching sequences in miRBase release 9.1. These miRNAs are almost exclusively 20–22 nt in length in both species. (C) rRNA as identified by overlap with annotated rRNA genes in the rice genome. The relative lack of ribosomal fragments below 18 nt likely corresponds to the cutoff imposed by gel purification. (D) tRNA as annotated by tRNAscan-SE. (E) Un-annotated, small RNAs not overlapping with sequences annotated by the aforementioned methods. Again, a peak is observed at 24 nt for P. contorta sequences, revealing a higher proportion of evolutionarily conserved sequences in this class reside in the 24-nt fraction than in the 23- or 25-nt fractions.