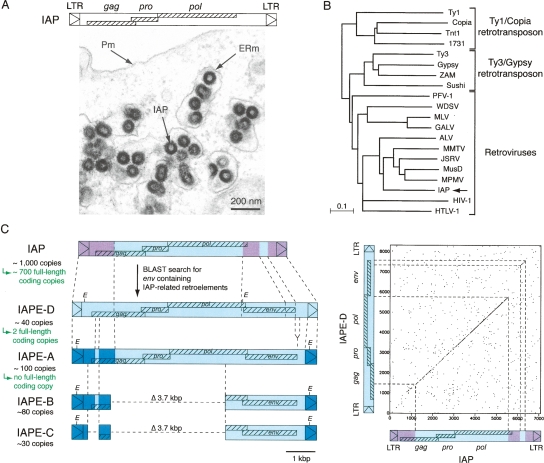
Figure 1.
Structure of IAPs and related elements. (A, top) Genomic organization of an IAP LTR-retrotransposon, with the LTRs flanking the three ORFs homologous to the retroviral gag, pro, and pol genes. (Bottom) Electron microscopy of 293T cells transfected with an expression vector for the retrotransposition competent IAP(92L23) copy, disclosing numerous typical IAP VLPs accumulated in the cisternae of the ER. (Pm) Plasma membrane; (ERm) endoplasmic reticulum membrane. (B) Phylogeny of LTR-retroelements, based on their reverse transcriptase (RT) domain. The tree was determined by the neighbor-joining method using the seven blocks of conserved residues found in the RT domain of all retroelements and was rooted using non-LTR retrotransposons. The retroviruses listed include alpha- (ALV), beta- (MMTV, JSRV, MPMV), gamma- (MLV, GALV), delta- (HTLV-1), epsilon- (WDSV) retroviruses; lentiviruses (HIV-1); and foamyviruses (PFV-1). All sequences are readily accessible from GenBank and previous reports (e.g., Malik and Eickbush 2001). (C) Search for IAP-related retroelements containing a coding-competent env gene. BLAST search in the C57BL/6 mouse genome identifies the already known IAPE-A, -B, and -C elements, together with the yet-unreported IAPE-D subfamily. Sequences of the representative IAPE-D element and the homologous conserved domains in other IAP and IAPE elements are indicated in light blue; non homologous domains between IAP and IAPE elements are indicated in purple; non homologous domains between IAPE-D and the other IAPE subfamily members are indicated in dark blue. The limits of the conserved domains between IAPs and IAPEs were determined by DotPlot analysis (see IAP/IAPE-D dot plot analysis on the right; window size = 15, threshold = 45). For each subfamily, the number of copies in the C57BL/6 genome is indicated together with the number of full-length coding copies.