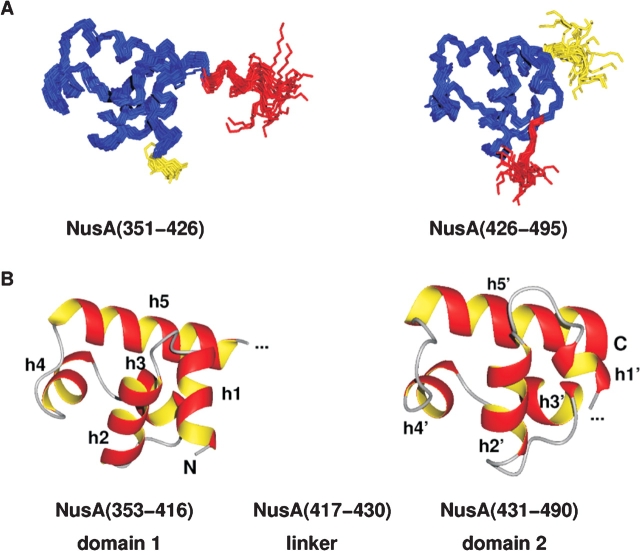
Figure 2.
Structure of the carboxy-terminal domain of NusA. (A) Backbone overlay of the 19 accepted structures for the two subdomains of the carboxy-terminal domain of NusA, NusA(351–426), and NusA(426–495), respectively. The linker residues are shown in red; flexible residues at the amino and carboxyl terminus are depicted in yellow. (B) Lowest energy structures of the relatively rigid parts of NusACTD, NusA(353–416), and NusA(431–490) in ribbon representation. Each subdomain contains two HhH motifs that are linked together by a so-called connector helix, thus adopting a compact (HhH)2 fold. Helices h1/h1′ and h2/h2′ constitute the first and h4/h4′ and h5/h5′ the second HhH motif, whereas h3/h3′ represent the connector helices. The linker between the two subdomains seems to be structured (see A, red part of overlay); however, 15N relaxation data indicate that the interdomain region comprising about 14 amino acids is flexible. The picture was drawn with MOLMOL (Koradi et al. 1996).