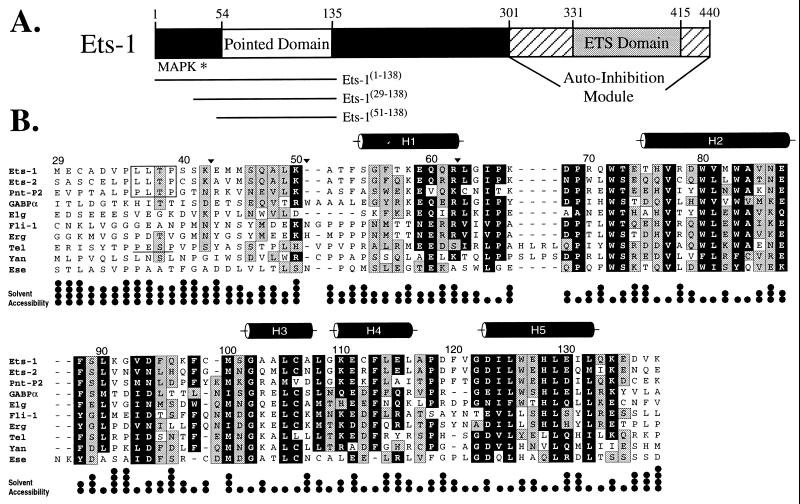
Figure 1.
The PNT domain is defined by a region of sequence conservation found in a subset of ets proteins. (A) Schematic diagram of the murine Ets-1 protein showing the locations of the PNT domain, MAP kinase phosphorylation site (Thr-38 ∗), and DNA-binding ETS domain with flanking autoinhibitory sequences (1). The central region of the protein contains putative transactivation domain(s). (B) Alignment of the sequences of PNT domains and preceding N-terminal regions from the ets family members murine Ets-1, Ets-2, GABPα, and Fli-1, human Erg, Tel, and Ese, and Drosophila PNT-P2, Elg, and Yan. The positions of highly conserved amino acids are highlighted in black (seven or more members having blosum62 substitution scores ≥1), and those of moderately conserved residues in gray (six or more members having blosum62 substitution scores ≥0). The MAP kinase phosphorylation sites identified in Ets-1, Ets-2, and PNT-P2 are boxed. Based on a consensus MAP kinase substrate sequence P-X-T/S-P, Tel also contains a potential phosphorylation site (underlined). Positions of tryptic cleavage in Ets-1(29–138) under conditions of partial proteolysis are denoted by ▾. The five α-helices (cylinders) in the Ets-1 PNT domain were identified by NMR methods. The fractional solvent accessibilities of the side chains in a low-energy structure of Ets-1(29–138) are illustrated (one • = 0–25%).