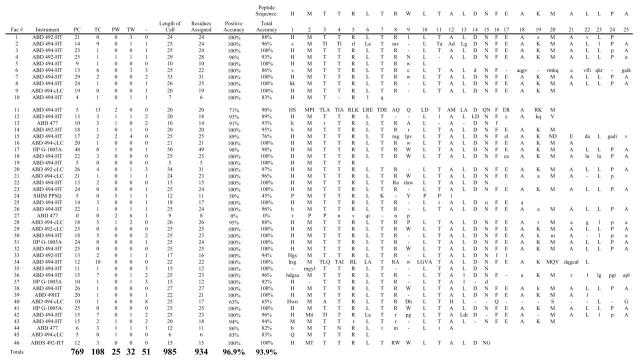
TABLE 1.
ESRG’03 Sequence Assignments from Facilities
Facility number (Fac#), Positive Correct (PC). Tentative Correct (TC), Positive Wrong (PW). Tentative Wrong (TW), No call (-). Uppercase letters represent positive calls and lowercase letters represent tentative assignments. Assignments where multiple positive calls were made with the correct amino acid were counted as a tentative correct Assignments where multiple positive calls were made without the correct assignment were counted as a positive wrong. Positive accuracy = PC/(PC + PW), total accuracy = (PC + TC)/(PC + TC + PW + TW).
Company abbreviations: AppliedBiosystems (ABD), Hewlett Packard/Agilent (HP), Shimadzu (SHIM).
Expected sequence past residue 2S:VRRYGRLTRATGLVLEATGLQLPLG.
A total of 7 facilities reported past 25 residues and the data were as follows: Facility #4, VsRY; Facility #7,V-RYg-LT; Facility #8,V; Facility #17.VRRYGRLTRATGLVLEATGLQ-PVG; Facility #20, V-rYg-ltl; Facility #28, v; Facility #38,VR. Facility #41 apparently called data past 25 based by circling pmol on the amino acid/pmol table submitted for repetitive yield calculations; VRRYGRL-TRATglVIE-TglQ-p.