Abstract
The structure of a novel c7-type cytochrome domain that has two bishistidine coordinated hemes and one heme with histidine, methionine coordination (where the sixth ligand is a methionine residue) was determined at 1.7 Å resolution. This domain is a representative of domains that form three polymers encoded by the Geobacter sulfurreducens genome. Two of these polymers consist of four and one protein of nine c7-type domains with a total of 12 and 27 hemes, respectively. Four individual domains (termed A, B, C, and D) from one such multiheme cytochrome c (ORF03300) were cloned and expressed in Escherichia coli. The domain C produced diffraction quality crystals from 2.4 M sodium malonate (pH 7). The structure was solved by MAD method and refined to an R-factor of 19.5% and R-free of 21.8%. Unlike the two c7 molecules with known structures, one from G. sulfurreducens (PpcA) and one from Desulfuromonas acetoxidans where all three hemes are bishistidine coordinated, this domain contains a heme which is coordinated by a methionine and a histidine residue. As a result, the corresponding heme could have a higher potential than the other two hemes. The apparent midpoint reduction potential, Eapp, of domain C is −105 mV, 50 mV higher than that of PpcA.
Keywords: multiheme cytochrome c, cytochrome c7, Geobacter metallireducens, Geobacter sulfurreducens, heme coordination in c-type cytochromes, protein structure
c-Type cytochromes have hemes covalently bound to the protein through two thioether bonds with cysteine residues; the largest family consist of the class I molecules (Mathews 1985; Degtyarenko et al. 1997 [http://metallo.scripps.edu/PROMISE/]), where the fifth and sixth ligands of the hemes are histidine and methionine residues, respectively. Bishistidine coordination is found in multiheme cytochromes (Degtyarenko et al. 1997 [http://metallo.scripps.edu/PROMISE/]): for example, class III (Coutinho and Xavier 1994), and class IV cytochromes (Deisenhofer et al. 1995), and in cytochrome c nitrite reductase (Einsle et al. 2000), and other four heme cytochromes (Leys et al. 2002). Only in class IV, the photosynthetic reaction center cytochrome c subunit, hemes of both coordination types are found in the same domain (Deisenhofer et al. 1995). Cytochrome c peroxidase has both a bishistidine coordinated heme and a heme with His–Met coordination; however, these hemes are in separate domains (Fülöp et al. 1995; Shimizu et al. 2001). In general, hemes with His–Met coordination are expected to have more positive reduction potentials than hemes with His–His coordination (for review, see Dolla et al. 1994).
Among the multiheme cytochromes from the class III cytochrome family, the cytochromes c3 from the sulfur- and sulfate-reducing bacteria are best studied (for review, see Mathews 1985; Coutinho and Xavier 1994; Aragao et al. 2003). They consist of about 120 amino acid residues and four covalently linked heme groups. The hemes are numbered sequentially according to the cysteine residues to which they are attached in the amino acid sequence. Heme IV is suggested to interact with hydrogenases (Aubert et al. 1997; Brugna et al. 1998). Although the sequence identity among the four-heme proteins is relatively low, ~25%, the positions and orientations of the hemes in the three-dimensional structures are maintained (Aragao et al. 2003). The three-heme cytochromes c7 from Fe(III)-reducing bacteria (Seeliger et al. 1998; Afkar and Fukumori 1999; Lloyd et al. 2003) are structurally homologous to the four-heme cytochromes c3, but are missing heme II of the cytochrome c3 molecules and the amino acid chain segment that keeps that heme in place. For comparison with the more extensively studied four-heme cytochromes, the cytochrome c3 nomenclature for labeling the hemes in the cytochrome c7 proteins is maintained.
We have previously expressed in Escherichia coli and determined the structure of cytochrome c7 from Geobacter sulfurreducens, designated PpcA, a protein with 71 residues that contains three covalently bound hemes (Londer et al. 2002; Lloyd et al. 2003; Pokkuluri et al. 2004). It is one of the smallest cytochrome c-type molecules with the highest ratio of hemes to protein residues. Searching the G. sulfurreducens genome with the PpcA sequence we identified three ORFs coding for proteins that are polymers formed with repeats of homologous c7-type domains. Two of the proteins (coded by ORF00991 and ORF03300) have four repeats (total of 12 hemes) and one (coded by ORF03649) has nine repeats (total of 27 hemes) of the cytochrome c7-type domain (Pokkuluri et al. 2004). We also found homologous polymers in the G. metallireducens genome (four unit polymers are encoded by Gmet_6, Gmet_8 and the nine unit polymer is encoded by Gmet_2 in the G. metallireducens genome [http://www.ncbi.nlm.nih.gov, accession number NZ_AAAS00000000]) with 77%, 75%, and 72% identity, respectively, with the ones identified in the G. sulfurreducens genome. We predicted based on the crystal packing observed in the PpcA crystals that the domains in the above polymers will be linearly arranged (Pokkuluri et al. 2004). The functions of the polymers are presently unknown. The repeats within each of the three poly-cytochrome c7-type proteins are highly homologous. The repeat lengths are longer than that of the cytochrome c7 PpcA molecule (71 residues); they vary from 73 to 82 residues.
These proteins appear to represent a new type of cytochrome. Interestingly, in each repeat of the multi-cytochrome c7 sequences, the sixth His ligand to heme IV is absent, the homologous residue in the sequence is mostly Lys or Gln. We proposed earlier that either the Lys or Gln residues or one of the two conserved Met residues in the vicinity could be the second axial ligand to heme IV in those domains (Pokkuluri et al. 2004). Because the chain segments between the heme I and heme III binding sites are longer in the poly-cytochrome c7-type domains than in the cytochrome c7 molecule PpcA, the structure of this segment, and therefore the residue that forms the sixth ligand of heme IV, could not be predicted with confidence.
To characterize these cytochrome c7-type domains, we have expressed in E. coli the four individual domains of the mature protein coded by ORF003300 (Fig. 1 ▶) of the G. sulfurreducens genome, and have determined the three-dimensional structure by X-ray diffraction of single crystals at 1.7 Å resolution of the third domain, domain C. In this manuscript, we describe the structural features of this protein in comparison with the structure of cytochrome c7 (PpcA) from G. sulfurreducens. The alignment of the amino acid sequence of domain C with that of PpcA is shown in Figure 2 ▶.
Figure 1.
Sequence of ORF03300 from G. sulfurreducens; its four domains are aligned with each other.
Figure 2.
Amino acid sequence alignment of domain C of the four domain poly-c7-type cytochrome (ORF03300) and cytochrome c7 (PpcA) from G. sulfurreducens. Identical residues are shown in bold; the heme binding residues are shown in gray boxes and the heme numbering is indicated below. The sixth axial ligand to heme IV in both proteins is shown in a black box (note that these residues do not align in the sequence).
Results and Discussion
Cloning and expression of the domains
For cloning the single domains, the expression system described by Londer et al. (2002) was modified as follows: (1) Genes were cloned without adding extra N-terminal residues to their mature sequences; (2) the gene coding for periplasmic chaperone, Skp, known to facilitate folding of various proteins in the periplasm (Bothmann and Pluckthun 1998; Schäfer et al. 1999; Mavrangelos et al. 2001) was cloned immediately after the ampicillin-resistance gene to be expressed constitutively. The new vector is named pVA203 (its detailed description will be published elsewhere).
We cloned separately each of the four cytochrome c7-type domains (A, B, C, and D) of the four-unit polymer (ORF03300) into the periplasmic expression vector pVA203. Domain A was cloned both as a fusion to the OmpA leader sequence and with its native leader sequence. All five constructs produced c-type cytochromes as judged by the pink color of the harvested cells. The overall yields of domains increased in the following order: A with native leader sequence << A with OmpA leader sequence < B < C << D. The yield of domain A was very low, and its isolation was not attempted. Domains B, C, and D were purified by cation-exchange chromatography and gel filtration as described previously (Londer et al. 2002). The yields of pure proteins per liter of cell culture were: domain B, 0.2 mg; domain C, 0.25 mg; domain D, 1.1 mg.
Structure determination
Crystals were obtained from domains C and D; crystals of domain C diffracted well and allowed structure determination at a resolution of 1.7 Å. The structure was solved by multiple wavelength anomalous dispersion (MAD) method with program CNS (Brünger et al. 1998) using data collected at the Fe K-absoption edge at the SBC beam line 19BM (APS). The structure was refined to 1.7 Å resolution with data collected at beam line 19ID (APS); the R-factor and R-free are 19.5% and 21.8%, respectively (see Table 1).
Table 1.
Summary of data collection and crystallographic parameters
| Crystal parameters | |||||
| Unit cell (Å) a = b = 43.8, c = 123.8 | |||||
| Space group P41212 | |||||
| MAD data | |||||
| Inflection | Peak | High remote | Low remote (reference) | High res | |
| Data collection | |||||
| Wavelength (Å) | 1.74044 | 1.73769 | 1.69031 | 1.78780 | 1.03320 |
| Resolution range (Å) | 99.0–2.20 | 99.0–2.15 | 99.0–2.05 | 99.0–2.20 | 99.0–1.7 |
| No. of unique reflections | 6628 | 7072 | 8024 | 6594 | 13,958 |
| Redundancy (last shell) | 9.5 (3.4) | 9.3 (3.5) | 9.0 (2.6) | 9.2 (3.4) | 10 (11) |
| Completeness (last shell), % | 99 (87) | 99 (89) | 97 (77) | 98 (88) | 99 (100) |
| Average I/σ(l) (last shell) | 28 (2.6) | 56 (9.4) | 45 (4.2) | 64 (9.2) | 61 (13) |
| R-merge (last shell), % | 8.7 (34) | 6.8 (15.2) | 6.8 (24.2) | 4.1 (14.7) | 5.0 (27.4) |
| Inflection | Peak | Remote | All | ||||
| Resolution range (50–2.4 Å) | Fried | Iso | Fried | Iso | Fried | Iso | |
| Phasing | |||||||
| Phasing power | 1.71 | 1.57 | 2.21 | 1.86 | 2.40 | 2.00 | |
| FOM | 0.45 | 0.44 | 0.46 | 0.44 | 0.37 | 0.36 | 0.81 |
| Density modification, FOM | 0.95 | ||||||
| a The test set for R-free calculation was comprised of 10% of the total reflections selected at random and were not used in refinement. | |
| Refinement | |
| Resolution Range (Å) | 99.0–1.7 |
| σcut-off used in refinement | 0.0 |
| No. of reflections | 24,246 |
| No. of nonhydrogen atoms refined (Average B-factor, Å2) | |
| Protein | 589 (24.6) |
| Heme | 129 (19.0) |
| Water | 105 (38.1) |
| R-factor, R-freea (all data) | 0.195, 0.218 |
| RMSD from ideal geometry | |
| Bond length (Å) | 0.008 |
| Angle (°) | 3.6 |
Comparison of domain C and PpcA
The structure of domain C clearly shows that the sixth ligand of heme IV is a methionine; we could not determine the identity of the coordinating residue based on the amino acid sequence alone. Domain C contains 82 amino acids, while PpcA has 71 amino acids. Although domain C also contains three hemes and similar number of amino acid residues its structure is different from that of the PpcA determined earlier in our laboratory (Figs. 3 ▶,4 ▶). Domain C, as cloned, has five additional residues at the N terminus compared to PpcA. The sequence alignment of the domains of ORF03300 shown in Figure 1 ▶ suggested that the N-terminal Lys and Gly residues of the cloned domain C should be at the end of domain B; however, they were included at the begining of domain C for proper periplasmic cleavage. Twenty-one out of 82 residues are identical between domain C and PpcA (Fig. 2 ▶), but only 16 residues have the same role in the protein structure; 11 of these residues bind the hemes. There is one deletion in domain C compared to PpcA; the equivalent residue of Lys 22 or Ala23 in PpcA is not present in domain C. There is an insertion of a Gly residue in the tip of the β-bend of domain C; four hydrophobic amino acids, Leu, Phe, Ala, and Tyr are inserted after the conserved lysine, Lys33 in PpcA; there is also an insertion of two residues (Asn and Gly) after the His residue of the heme III binding site.
Figure 3.
Ribbon drawing of domain C (left) and overlap of domain C and PpcA (right). Domain C and PpcA were overlapped using heme IV atoms. Domain C, blue; PpcA, magenta; hemes in domain C, green; hemes in PpcA, gray.
Figure 4.
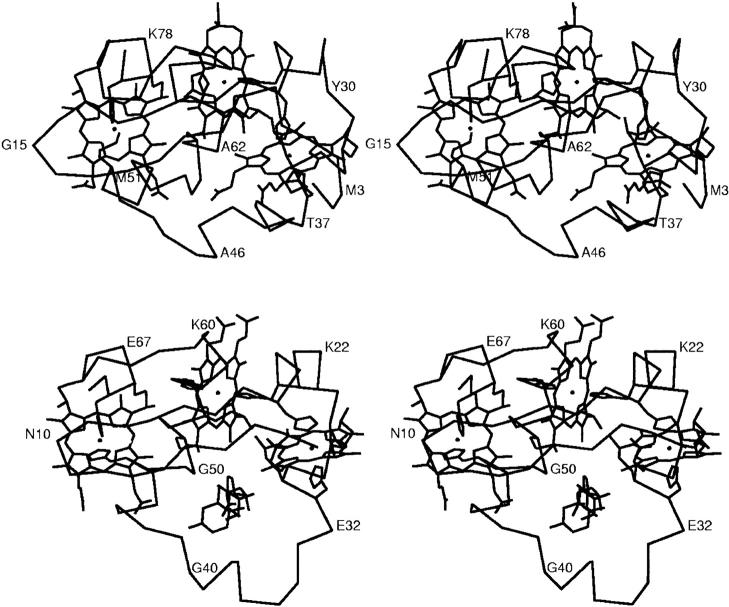
Stereo views showing hemes, α carbons, and side chains that bind the hemes in (top) domain C, (bottom) cytochrome c7 (PpcA) from G. sulfurreducens. The PpcA structure shown also includes a molecule of deoxycholic acid in the lower part of the figure. Heme I is on the right, heme III is in the middle, and heme IV is located on the left side of the figures. The orientations of the hemes IV are the same in the two molecules. Note that the methionine residue that coordinates heme IV in domain C is in the left side of the figure.
As was observed in the PpcA structure, domain C also has a two-stranded β-sheet segment near its N terminus, and five helical regions, one less than PpcA. The helical segments include the three heme-binding sites, the segment that contain the sixth ligands to hemes I, III, and the segment that contains the sixth ligand to heme IV. Although the latter segments in the two structures are in homologous positions in the amino acid sequences (Fig. 2 ▶), their relative positioning in structures compared to heme IV are different. In the case of PpcA, His47, the sixth ligand to heme IV is located at the end of the helical segment, whereas Met51, which is the sixth ligand to heme IV in domain C, is at the beginning of the helical segment. The rotamer conformation of the Met51 is chi1 -66, chi2 -50, chi3 -49. Lovell et al. (2000) found that this rotamer is the most observed in their data base; it is also one of the most observed for methionines in helical regions. Interestingly, this rotamer is not used by any of the methionines that are ligands to hemes in other cytochrome c proteins (P.R. Pokkuluri and M. Schiffer, unpubl.).
Domain C has a more conventional structure than PpcA. The number of hydrophobic residues larger than Ala in the core of domain C and the number of side-chain-to-main-chain hydrogen bonds (see Table 2) are more similar to what is observed for globular proteins in general; they are very low in PpcA. Domain C has 12 Lys and one Arg residue, compared with 17 Lys and no Arg residues in PpcA.
Table 2.
Hydrogen bonds formed by heme propionic acid groups and by His residues
| Hydrogen bonded atoms | Distance (Å) | |
| Heme I O2A | Ala44 N | 2.85 |
| Heme I O1D | Gly45 N | 2.93 |
| Heme III O2D | Phe70 N | 2.93 |
| Heme III O2D | Arg48 NH1 | 2.61 |
| Heme IV O2A | Met51 N | 2.76 |
| Heme IV O1D | Phe49 N | 2.88 |
| His23 ND1 | Pro6 O | 2.75 |
| His26 ND1 | Ser22 O | 2.87 |
| His36 ND1 | Phe40 O | 2.89 |
| His36 ND1 | Ala41 O | 3.16 |
| His64 ND1 | Phe70 O | 2.90 |
| His80 ND1 | Ala17 O | 2.78 |
The ND1 atoms of all five of the histidine residues are hydrogen bonded to main-chain carbonyl oxygen atoms. Due to the differences in the structures, only two histidine ND1 atoms, from His 22 and His80, are hydrogen bonded to equivalent carbonyls in PpcA. In contrast to PpcA, none of the histidine residues in domain C are accessible to solvent. There is an unusual cis-peptide bond between His36 and Thr37 (Fig. 5 ▶). Nonproline cis-peptide bonds are very rare; they represent only 0.03% of the peptide bonds in PDB (Jabs et al. 1999). The conformation of the chain segment that consists of hydrophobic residues Leu39, Phe40, Ala41, and Tyr42 is probably made possible by the presence of the above cis-peptide bond in domain C. These residues shield heme I, the least exposed heme in this structure, from solvent. The ring of Phe40 is close to heme I; carbonyls of residues Phe40 and Ala41 are hydrogen bonded to the ND1 atom of His36, the fifth ligand to heme I. The ring of Tyr42 is near and close to parallel to His36.
Figure 5.
The cis-peptide bond observed in domain C between residues His-36 and Thr-37. Electron density in the final simulated-annealing composite omit-map generated by the program CNS is contoured at 1σ.
All but one of the heme propionic acids form hydrogen bonds with a backbone amide nitrogen; in addition, a salt bridge is formed between Arg48 and propionic acid D of heme III. Heme III has the largest accessible surface area of 173 Å2, followed by heme IV of 167 Å2, and heme I is least exposed with 79 Å2. In PpcA, heme I had the largest accessible surface area of 225 Å2 and heme III had the smallest one of 144 Å2. In the full-length protein, we predict that only heme III would be exposed to the solution, whereas both hemes I and IV would be largely buried in the interface of neighboring domains. This is expected, as we predict that the four domains coded by ORF03300 will be linearly arranged in the full-length protein.
The iron to iron distances of the hemes III and IV, hemes III and I, and hemes IV and I, respectively, are 11.4, 12.0, and 17.8 Å in domain C compared to 11.2, 12.6, and 20.8 Å in PpcA. These Fe–Fe distances are probably determined by the differences in number of residues between the heme binding sites. Surprisingly, the iron to iron distances found in this c7-type domain are closer to the ones observed in cytochromes c3 (average Fe–Fe distances between hemes III and IV, hemes III and I, and hemes I and IV for six proteins are 11.1, 12.2, and 17.7 Å (Higuchi et al. 1984; Matias et al. 1993, 1996; Czjzek et al. 1994; Morais et al. 1995; Einsle et al. 2001).
Crystal packing
As we previously observed in the PpcA crystal packing, the domain C molecules also form a long chain in the crystal. In domain C crystal, molecules are arranged in infinite chains parallel to the crystallographic c axis with the neighboring molecules related by a 41 axis (90° rotation and 1/4 unit cell translation) instead of one unit-cell translations along a and b axes as in the PpcA crystal (Pokkuluri et al 2004). In both crystals, the molecules in these chains are arranged such a way that heme I from one molecule is close to heme IV of the neighboring molecule. The iron to iron distance between these hemes in neighboring molecules in domain C is 13.8 Å compared to 12.9 Å in PpcA, but due to the difference in the relative angles of the prophyrin rings the heme edges are closer together in domain C crystals with the CMB atoms of heme I and heme IV 3.9 Å apart compared to 5.6 Å between the same atoms in PpcA crystals. Although the molecules also form infinite chains in the crystal of domain C, the N-and C-terminii cannot be connected, as they point in the opposite directions, and therefore, the crystal packing in domain C cannot form a model for the polymers, whereas the crystal packing in PpcA clearly suggested a model for them.
Visible redox titrations
The redox behavior of domain C and PpcA was investigated by redox titrations followed by visible spectroscopy. Results are given in Figure 6 ▶. The analysis of the redox titrations of the two proteins showed that the curve corresponding to domain C is clearly shifted to more positive reduction potential values relative to that of PpcA. This shift is reflected in the apparent midpoint reduction potential, Eapp, of each protein (i.e., the point at which the oxidized and reduced fractions are equal). The value of Eapp for PpcA is −155 mV; it is increased by 50 mV to −105 mV for domain C. The observed positive direction of the shift in the redox curves was expected (Mus-Veteau et al. 1992; Dolla et al. 1994) on the basis of different heme IV axial coordination for PpcA (His–His) and domain C (His–Met).
Figure 6.
Visible redox titrations for domain C (open squares) and PpcA (open circles) at 298 K (pH 7.9). The molar fraction of the total reduced protein was fitted to the model described by Santos et al. (1984) and Wang et al. (1991), for various solution potentials. Solid lines indicate the results of the fits for the three macroscopic reduction potentials (vs. standard hydrogen electrode), which in domain C are −185 mV, −98 mV, and -52 mV, and in PpcA are −200 mV, −155 mV, and −110 mV.
In summary, we have determined the structure of domain C of the four domain protein coded by ORF03300 of G. sulfurreducens. The structure has shown that the sixth ligand to heme IV is a methionine. This could not have been predicted based on sequence homology to three-heme cytochromes c7. There is a conserved methionine at equivalent position in the sequences of all polymers made up of c7-type domains (like domain C) found in G. sulfurreducens (Pokkuluri et al 2004) and G. metallireducens, suggesting that the sixth ligand to heme IV in all domains is a methionine. These domains represent a new type of multiheme cytochrome c as they have two hemes with bis-His coordination and one with His–Met coordination. As expected, due to this difference in heme coordination, the apparent midpoint reduction potential of this domain is more positive compared to the cytochrome c7 (PpcA).
Materials and methods
Cloning, expression, and purification of the domains
Cloning of the domains was carried out with modification of the methods previously described (Londer et al. 2002; Pokkuluri et al. 2004). A NotI restriction site was introduced at the end of the DNA fragment coding for the OmpA leader sequence of expression vector pCK32 (Londer et al. 2002). Mutagenesis resulted in Gln to Ala mutation in position “−2”, a position that is considered nonessential for proper processing of a leader sequence during secretion (Nielsen et al. 1997). This modification allows cloning genes without adding extra N-terminal residues to their mature sequences. Further, a unique NcoI restriction site was created 8 bp downstream of the ampicillin resistance gene, and the gene coding for E. coli periplasmic chaperone, Skp, was cloned into this site. The new vector was named pVA203. DNA fragments coding for the putative domains were amplified from G. sulfurreducens genomic DNA and cloned into the vector pVA203. Domain A was cloned both as a fusion to the OmpA leader sequence and together with its native leader sequence. Compared to the alignment shown in Figure 1 ▶, one N-terminal residue was removed in domains B and D, and two preceding residues (Lys–Gly) were added to the N terminus of domain C to ensure proper cleavage of the modified OmpA leader sequence as predicted by the SignalP software (www.cbs.dtu.dk/services/SignalP; Nielsen et al. 1997).
Expression and purification of the domains were performed with slight modification of the procedure previously described (Londer et al. 2002); induction was carried out with 10 μM IPTG and a final gelfiltration step was added to the purification protocol.
Crystallization of domain C
The protein was crystallized by hanging drops from 2.4 M sodium malonate (pH 7) (Hampton Research, Index screen-27). Tetragonal crystals, space group P41212 or P43212, with a unit cell of a = b = 43.8 Å, c = 123.8 Å, diffracted to a resolution of about 1.7 Å at the Structural Biology Center’s 19ID beam line at the Advanced Photon Source (APS).
Structure determination of domain C
Structure of domain C was determined by MAD method using four wavelength data sets (see Table 1) collected at the Fe K absorption edge at the 19BM (APS). Crystals were flash-cooled by briefly transferring them from mother liquor to a cryoprotectant solution (Index screen-27 containing 25% ethylene glycol). Data were processed with HKL2000 (Otwinowski and Minor 1997). Data collected at low-energy remote wavelength was used as the reference data set for structure determination using MAD method. Structure was solved with the program CNS (Brünger et al. 1998). After density modification the electron density map generated in space group P41212 was of excellent quality (FOM = 0.95) and allowed model building. The protein model was built manually into the electron density map using the program CHAIN (Sack 1988) and refined against high resolution data collected at 19ID (APS). Refinement of the model was carried out with CNS (Brünger et al. 1998). There are no residues in the disallowed regions of the Ramachandran plot, and 87% residues are in most favored regions. The data collection and refinement parameters are summarized in Table 1. The structure and structure factors were deposited in the Protein Data Bank with accession code, 1rwj. Figure 3 ▶ was generated by the program SETOR (Evans 1993); Figures 4 ▶ and 5 ▶ were generated by the program CHAIN (Sack 1988). Accessible surface area was calculated by the program SURFACE (Lee and Richards 1971).
Sequence database search and analysis
Protein sequences homologous to the cytochrome c7 were identified by searching the G. sulfurreducens and G. metallireducens genomes (www.ncbi.nlm.nih.gov, accession numbers NC_002939 and NZ_AAAS00000000) with the sequence of the cytochrome c7 (PpcA, ORF01023) using BLAST software (Altschul et al. 1997). The SignalP software (http://www.cbs.dtu.dk/services/SignalP; Nielsen et al. 1997) was used for prediction of leader sequence cleavage sites. Genetic Computer Group Software package (Genetics Computer Group) was used for alignments.
Redox titrations followed by visible spectroscopy
Anaerobic redox titrations of domain C and PpcA, followed by visible spectroscopy were performed as described by Louro et al. (2001), with approximately 18 μM protein solutions in 100 mM Tris-maleate buffer at pH 7.9 and 298 ± 1 K. To check for hysteresis, each redox titration was performed in both oxidative and reductive directions, using sodium dithionite as the reductant, and potassium ferricyanide as the oxidant. To ensure a good equilibrium between the redox centers and the working electrode (Dutton 1978), a mixture of the following redox mediators were added to domain C and PpcA solutions, all at approximately 4 μM final concentration: Methylene Blue, gallocyanine, Indigo Tetrasulphonate, Indigo Trisulphonate, Indigo Disulphonate, anthraquinone-2,7-disulphonate, 2-hydroxy-1,4-naphthoquinone, anthraquinone-2-sulphonate, safranine O, diquat, benzylviologen, Neutral Red, methylviologen. For domain C the following additional mediators were also used, all at approximately 4 μM final concentration: p-benzoquinone, 1,2-naphtoquinone-4-sulfonic acid, 1,2-naphtoquinone, trimethylhydroquinone, phenasine methosulphate, and phenasine ethosulphate. The reduced fraction of each protein was determined using the α band peak, 552 nm for domain C and 551 nm for PpcA, according to the method described by Louro et al. (2001).
Acknowledgments
This work is supported by the U.S. Department of Energy’s Office of Science, Biological and Environmental Research, Structural Biology, and NABIR programs under contract No. W-31-109-Eng-38. Use of the Argonne National Laboratory Structural Biology Center beam lines at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-310109-Eng-38. We thank Dr. Randy Alkire for help with data collection at the Fe K edge. Miguel Pessanha acknowledges Fundação para a Ciência e Tecnologia (FCT) for a doctoral fellowship (SFRH/5229/2001).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
Eapp, apparent midpoint reduction potential
MAD, multiple wavelength anamolous dispersion
PDB, Protein Data Bank
Article published online ahead of print. Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04626204.
References
- Afkar, E. and Fukumori, Y. 1999. Purification and characterization of triheme cytochrome c7 from the metal-reducing bacterium, Geobacter metallireducens. FEMS Microbiol. Lett. 175 205–210. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragao, D., Frazao, C., Sieker, L., Sheldrick, G.M., LeGall, J., and Carrondo, M.A. 2003. Structure of dimeric cytochrome c3 from Desulfovibrio gigas at 1.2 Å resolution. Acta Crystallogr. D 59 644–653. [DOI] [PubMed] [Google Scholar]
- Aubert, C., Leroy, G., Bruschi, M., Wall, J.D., and Dolla, A. 1997. A single mutation in the heme 4 environment of Desulfovibrio desulfuricans Norway cytochrome c3 (Mr 26,000) greatly affects the molecule reactivity. J. Biol. Chem. 272 15128–15134. [DOI] [PubMed] [Google Scholar]
- Bothmann, H. and Pluckthun, A. 1998. Selection for a periplasmic factor improving phage display and functional periplasmic expression. Nat. Biotechnol. 16 376–380. [DOI] [PubMed] [Google Scholar]
- Brugna, M., Giudici-Orticoni, M.T., Spinelli, S., Brown, K., Tegoni, M., and Bruschi, M. 1998. Kinetics and interaction studies between cytochrome c3 and Fe-only hydrogenase from Desulfovibrio vulgaris Hildenborough. Proteins 33 590–600. [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., Delano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nigles, M., Pannu, N.S, et al. 1998. Crystallography and NMR system: A new software suite for macro-molecular structure determination. Acta Crystallogr. D 54 905–921. [DOI] [PubMed] [Google Scholar]
- Coutinho, I.B. and Xavier, A.V. 1994. Tetraheme cytochromes. Methods Enzymol. 243 119–140. [DOI] [PubMed] [Google Scholar]
- Czjzek, M., Payan, F., Guerlesquin, F., Bruschi, M., and Haser, R. 1994. Crystal structure of cytochrome c3 from Desulfovibrio desulfuricans Norway at 1•7 Å resolution. J. Mol. Biol. 243 653–667. [DOI] [PubMed] [Google Scholar]
- Degtyarenko, K.N., North, A.C.T., and Findlay, J.B.C. 1997. PROMISE: A new database of information on prosthetic centres and metal ions in protein active sites. Protein Eng. 10183–186. [DOI] [PubMed] [Google Scholar]
- Deisenhofer, J., Epp, O., Sinning, I., and Michel, H. 1995. Crystallographic refinement at 2.3 Å resolution and refined model of the photosynthetic reaction centre from Rhodopseudomonas viridis. J. Mol. Biol. 246 429–457. [DOI] [PubMed] [Google Scholar]
- Dolla, A., Blanchard, L., Guerlesquin, F., and Bruschi M. 1994. The protein moiety modulates the redox potential in cytochromes c. Biochimie 76 471–479. [DOI] [PubMed] [Google Scholar]
- Dutton, P.L. 1978. Redox potentiometry: Determination of mid-point potentials of oxidation-reduction components of biological electron-transfer systems. Methods Enzymol. 54 411–435. [DOI] [PubMed] [Google Scholar]
- Einsle, O., Stach, P., Messerschmidt A., Simon, J., Kröger, A., Huber, R., and Kroneck, P.M.H. 2000. Cytochrome c nitrite reductase from Wolinella succinogenes. J. Biol. Chem. 275 39608–39616. [DOI] [PubMed] [Google Scholar]
- Einsle, O., Foerster, S., Mann, K., Fritz, G., Messerschmidt, A., and Kroneck, P.M.H. 2001. Spectroscopic investigation and determination of reactivity and structure of the tetraheme cytochrome c3 from Desulfovibrio desulfuricans Essex 6. Eur. J. Biochem. 268 3028–3035. [DOI] [PubMed] [Google Scholar]
- Evans, S.V. 1993. Hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graphics 11134–138. [DOI] [PubMed] [Google Scholar]
- Fülöp, V., Ridout, C.J., Greenwood, C., and Hajdu, J. 1995. Crystal structure of the dihaem cytochrome c peroxidase from Pseudomonas aeruginosa. Structure 3 1225–1233. [DOI] [PubMed] [Google Scholar]
- Higuchi, Y., Kusunoki, M., Matsuura, Y., Yasuoka, N., and Kakudo, M. 1984. Refined structure of cytochrome c3 at 1.8 Å resolution. J. Mol. Biol. 172 109–139. [DOI] [PubMed] [Google Scholar]
- Jabs, A., Weiss, M.S., and Hilgenfeld, R. 1999. Non-proline cis peptide bonds in proteins. J. Mol. Biol. 286 291–304. [DOI] [PubMed] [Google Scholar]
- Lee, B. and Richards, F.M. 1971. The interpretation of protein structures: Total volume, group volume distributions and packing density. J. Mol. Biol. 55 379–400. [DOI] [PubMed] [Google Scholar]
- Leys, D., Meyer, T.E., Tsapin, A.S., Nealson, K.H., Cusanovich, M.A., and Van Beeumen, J.J. 2002. Crystal structures at atomic resolution reveal the novel concept of “Electron-harvesting” as a role for the small tetraheme cytochrome c. J. Biol. Chem. 277 35703–35711. [DOI] [PubMed] [Google Scholar]
- Lloyd, J.R., Leang, C., Hodges Myerson, A.L., Coppi, M.V., Cuifo, S., Methe, B., Sandler, S.J., and Lovley, D.R. 2003. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londer, Y.Y., Pokkuluri, P.R., Tiede, D.M., and Schiffer, M. 2002. Production and preliminary characterization of a recombinant triheme cytochrome c7 from Geobacter sulfurreducens in Escherichia coli. Biochim. Biophys. Acta 1554 202–211. [DOI] [PubMed] [Google Scholar]
- Louro, R.O., Catarino, T., LeGall, J., Turner, D.L., and Xavier, A.V. 2001. Cooperativity between electrons and protons in a monomeric cytochrome c3: The importance of mechanochemical coupling for energy transduction. Chembiochem. 2 831–837. [DOI] [PubMed] [Google Scholar]
- Lovell, S.C., Word, J.M., Richardson, J.S., and Richardson, D.C. 2000. The penultimate rotamer library. Proteins 40 389–408. [PubMed] [Google Scholar]
- Mathews, F.S. 1985. The structure, function and evolution of cytochromes. Prog. Biophys. Mol. Biol. 45 1–56. [DOI] [PubMed] [Google Scholar]
- Matias, P.M., Frazão, C., Morais, J., Coll, M., and Carrondo, M.A. 1993. Structure analysis of cytochrome c3 from Desulfovibrio vulgaris Hildenborough at 1•9 Å resolution. J. Mol. Biol. 234 680–699. [DOI] [PubMed] [Google Scholar]
- Matias, P.M., Morais, J., Coelho, R., Carrondo, M.A., Wilson, K., Dauter, Z., and Sieker, L. 1996. Cytochrome c3 from Desulfovibrio gigas: Crystal structure at 1.8 Å resolution and evidence for a specific calcium-binding site. Protein Sci. 5 1342–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrangelos, C., Thiel, M., Adamson, P.J., Millard, D.J., Nobbs, S., Zola, H., and Nicholson, I.C. 2001. Increased yield and activity of soluble single-chain antibody fragments by combining high-level expression and the Skp periplasmic chaperonin. Protein Expr. Purif. 23 289–295. [DOI] [PubMed] [Google Scholar]
- Morais, J., Palma, P.N., Frazão, C., Caldeira, J., LeGall, J., Moura, I., Moura, J.J.G., and Carrondo, M.A. 1995. Structure of the tetraheme cytochrome from Desulfovibrio desulfuricans ATCC 27774: X-ray diffraction and electron paramagnetic resonance studies. Biochemistry 34 12830–12841. [DOI] [PubMed] [Google Scholar]
- Mus-Veteau, I., Dolla, A., Guerlesquin, F., Payan, F., Czjzek, M., Haser, R., Bianco, P., Haladjian, J., Rapp-Giles, B.J., and Wall, J.D. 1992. Site-directed mutagenesis of tetraheme cytochrome c3. Modification of oxidore-duction potentials after heme axial ligand replacement. J. Biol. Chem. 267 16851–16858. [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10 1–6. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Pokkuluri, P.R., Londer, Y.Y., Duke, N.E.C., Long, W.C., and Schiffer, M. 2004. Family of cytochrome c7-type proteins from Geobacter sulfurreducens: The structure of one cytochrome c7 at 1.45 Å resolution. Biochemistry 43 849–859. [DOI] [PubMed] [Google Scholar]
- Sack, J.S. 1988. CHAIN—A crystallographic modeling program. J. Mol. Graphics 6 224–225. [DOI] [PubMed] [Google Scholar]
- Santos, H., Moura, J.J.G., Moura, I., LeGall, J., and Xavier, A.V. 1984. NMR studies of electron transfer mechanisms in a protein with interacting redox centres: Desulfovibrio gigas cytochrome c3. Eur. J. Biochem. 141 283–296. [DOI] [PubMed] [Google Scholar]
- Schäfer, U., Beck, K., and Müller, M. 1999. Skp, a molecular chaperone of Gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 274 24567–24574. [DOI] [PubMed] [Google Scholar]
- Seeliger, S., Cord-Ruwisch, R., and Schink, B. 1998. A periplasmic and extra-cellular c-type cytochrome of Geobacter sulfurreducens acts as a ferric iron reductase and as an electron carrier to other acceptors or to partner bacteria. J. Bacteriol. 180 3686–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, H., Schuller, D.J., Lanzilotta, W.N., Sundaramoorthy, M., Arciero, D.M., Hooper, A.B., and Poulos, T.L. 2001. Crystal structure of Nitrosomonas europa cytochrome c peroxidase and the structural basis for ligand switching in bacterial diheme peroxidases. Biochemistry 40 13483–13490. [DOI] [PubMed] [Google Scholar]
- Wang, D.L., Stankovich, M.T., Eng, L.H., and Neujahr, H.Y. 1991. Redox properties of cytochrome c3 from Desulfovibrio desulfuricans NCIMB 8372. Effects of electrode materials and sodium chloride. J. Electroanal. Chem. 318 291–307. [Google Scholar]