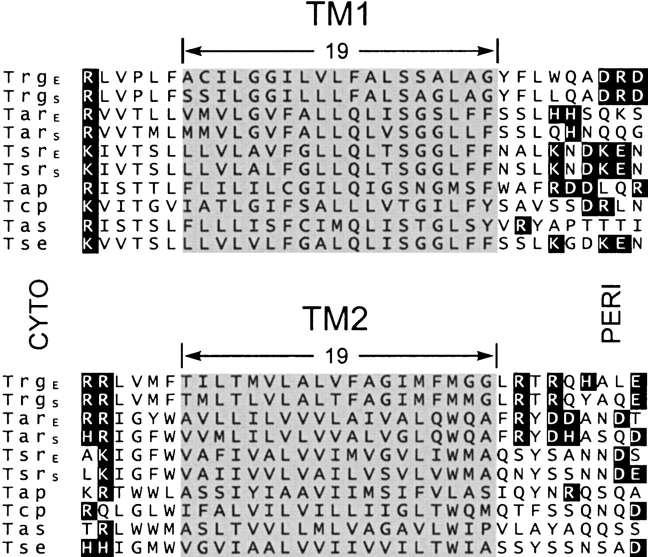
Figure 7.
Sequence alignments of TM1 and TM2 regions from enteric bacteria. The figure shows the TM1 and TM2 regions of a computer-generated alignment of the complete sequences of chemoreceptors Trg, Tar, and Tsr from E. coli and from Salmonella, Tap from E. coli, Tcp from Salmonella, and Tas and Tse from Enterobacter aerogenes. There are few residue identities in the regions shown. However, alignment in the TM2 region is the result of extension without gaps from the extensive stretch of residue identity among the sequences over much of the ~350-residue carboxyl terminal domain. Alignment in the TM1 region derives primarily from the correspondence of a cluster of positively charged residues on the amino-terminal side of the extended hydrophobic sequences. Charged residues are in dark boxes, and the segments identified as embedded in the hydrophobic environment for Trg are shaded for all sequences.