Figure 6.
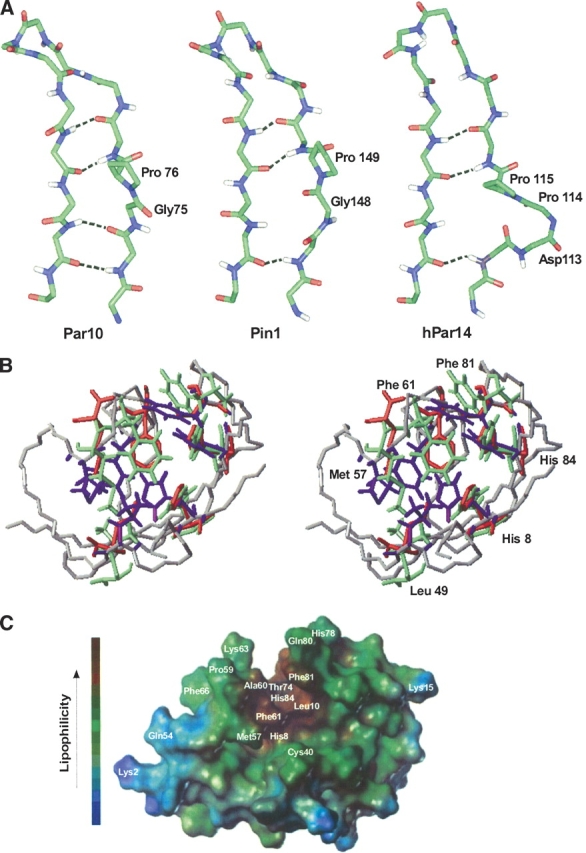
Orientation of the H-bonds in the β-sheet and putative substrate binding site of different Parvulins. (A) Arrangement of the G75-P76 cis amide bond in E. coli Par10, compared with the two homologous structures hPin1 and hPar14. In spite of the bulge resulting from the cis peptide bond, the β-sheet of E. coli Par10 can form all four H-bonds whereas the structures of hPar14 and hPin1 with trans-Pro are only stabilized by three H-bonds in this region (figure produced with Insight II, MSI Inc.). (B) Comparison of the putative binding pockets of hPar14 (PDB code 1eq3, green residues) and hPin1 (PDB code 1pin; red residues) overlaid on the E. coli Par10 structure (PDB code 1jnt; blue residues, grey backbone). The labeled residues are highly conserved in all parvulins and may be involved in the catalytic activity of these parvulins (figure produced with MOLMOL, version 2k.1; Koradi et al. 1996). (C) Connolly surface of E. coli Par10. Helix 4 (left) and the curved β-sheet form a lipophilic gap (brown), enabling a lipophilic oligopeptide substrate to bind (figure produced with the MOLCAD module of the program Sybyl, version 6.3; Tripos AG).
