Table 1.
Structural restraints and statistics for the NMR structures
| Restraints for structure calculation | ||
| Total NOE restraints | 723 | |
| Intraresidue | 275 | |
| Sequential | 162 | |
| Medium range (1 < |i - j| ≤ 5) | 89 | |
| Long range (|i - j| > 5) | 143 | |
| Hydrogen bonds | 54 | |
| Dihedral angles (3) | 39 | |
| Statistics for structure calculations | ||
| RMSD from distance restraints | ||
| NOE restraints (Å) | 0.016 ± 0.006 | |
| Dihedral restraints (°) | 0.46 ± 0.42 | |
| RMSD from idealized geometry | ||
| Bonds (Å) | 0.016 ± 0.003 | |
| Angle (°) | 0.35 ± 0.03 | |
| Impropers (°) | 0.19 ± 0.06 | |
| Mean energies (kcal/mole) | ||
| Etotal | 75.27 ± 19.50 | |
| Ebond | 3.89 ± 1.33 | |
| Eangle | 50.67 ± 8.78 | |
| Evdw | 7.81 ± 7.61 | |
| Enoc | 11.40 ± 7.26 | |
| Edihe | 0.91 ± 0.91 | |
| RMSD of atomic coordinates (Å) | Backbone | Heavy atoms |
〈SA〉ka vs. 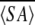 kr kr
| ||
| All residues | 1.195 | 1.907 |
| Secondary structures | 0.747 | 1.292 |
a 〈SA〉k is the ensemble of 20 final simulated annealing structures of MTH1880.
〈SA〉kr is the mean structure obtained by averaging the individual structures following a superimposition of the backbone heavy atoms.
