Abstract
Mammalian capping enzymes are bifunctional proteins with both RNA 5′-triphosphatase and guanylyltransferase activities. The N-terminal 237-aa triphosphatase domain contains (I/V)HCXXGXXR(S/T)G, a sequence corresponding to the conserved active-site motif in protein tyrosine phosphatases (PTPs). Analysis of point mutants of mouse RNA 5′-triphosphatase identified the motif Cys and Arg residues and an upstream Asp as required for activity. Like PTPs, this enzyme was inhibited by iodoacetate and VO43− and independent of Mg2+, providing additional evidence for phosphate removal from RNA 5′ ends by a PTP-like mechanism. The full-length, 597-aa mouse capping enzyme and the C-terminal guanylyltransferase fragment (residues 211–597), unlike the triphosphatase domain, bound poly (U) and were nuclear in transfected cells. RNA binding was increased by GTP, and a guanylylation-defective, active-site mutant was not affected. Ala substitution at positions required for the formation of the enzyme-GMP capping intermediate (R315, R530, K533, or N537) also eliminated poly (U) binding, while proteins with conservative substitutions at these sites retained binding but not guanylyltransferase activity. These results demonstrate that the guanylyltransferase domain of mammalian capping enzyme specifies nuclear localization and RNA binding. Association of capping enzyme with nascent transcripts may act in synergy with RNA polymerase II binding to ensure 5′ cap formation.
The 5′-terminal cap, m7GpppN, which is present on most eukaryotic mRNAs (1), plays crucial roles in enhancing mRNA processing (2–5), stability (6–9), nuclear transport (10, 11), and translation (1, 12–14). Cap formation is catalyzed by the cotranscriptional, sequential action of three enzymes. The γ-phosphate on the nascent RNA terminus is first removed by RNA 5′-triphosphatase (RTP) to generate a 5′ diphosphorylated end; GMP derived from GTP is transferred by RNA guanylyltransferase to the 5′ end to form a 5′-5′ triphosphate structure, GpppN-; and the guanine N7 position subsequently is methylated by RNA (guanine-7-) methyltransferase (1, 4, 15).
Although this pathway was identified more than 20 years ago (16, 17), cellular cap-forming enzymes have been cloned and sequenced only more recently (18–26). Capping in yeast species is mediated by separately encoded RTP and guanylyltransferase proteins (18), while higher eukaryotes contain bifunctional capping enzymes with N-terminal RTP and C-terminal guanylyltransferase (22, 27, 28). Despite this difference in genomic organization, capping function apparently has been conserved between unicellular and metazoan organisms. Yeast strains lacking RTP or guanylyltransferase, which are both essential, were complemented for growth by the mammalian counterparts (22, 23, 26).
In mammalian capping enzyme, the C-terminal guanylyltransferase domain contains several motifs characteristic of the nucleotidyl transferase superfamily (19, 22, 28, 29). These include the highly conserved KXDG motif, which is required for formation of the capping enzyme intermediate by phosphoamide linkage of GMP to the ɛ-amino group of the motif Lys (4). Ala substitution of this Lys eliminated guanylylation (22). The mammalian capping enzyme N-terminal 5′-triphosphatase domain also contains a conserved motif, (I/V)HCXXGXXR(S/T)G, which is characteristic of protein tyrosine phosphatases (PTPs) that form a motif cysteinyl-phosphate (Cys-P) intermediate as an essential part of their catalytic mechanism (30–32). This similarity was first described in Caenorhabditis elegans RTP where replacement of the motif Cys eliminated 5′-triphosphatase activity (27). This suggested a similar mechanism for Pi release from RNA 5′ termini and from protein phosphotyrosines, although it remained possible that the motif Cys was required for disulfide-mediated correct folding rather than as an active-site phosphate acceptor.
Recently, it was found that mammalian capping enzyme interacts with the elongating, hyperphosphorylated form of RNA polymerase II (pol IIo) (22, 33, 34), thus providing an explanation for the selective capping of transcripts made by pol II. Vaccinia virus capping enzyme also forms specific complexes with the viral RNA polymerase (35) and binds RNA as well (36). A general mechanism for RNA guanylylation, proposed on the basis of the crystal structure of Chlorella virus PBCV-1 capping enzyme (37), suggested direct binding to target RNAs. In addition, it was shown recently that Yersinia PTP can bind RNA and that binding is dependent on the conserved motif Cys (32).
In this report we demonstrate that full-length mouse capping enzyme (MCE) and the C-terminal guanylyltransferase fragment, but not the RTP domain, are localized to nuclei and bind RNA directly. In addition, the sites in MCE that are required for RTP activity indicate that a common mechanism is used by RTPs and PTPs.
MATERIALS AND METHODS
Plasmids.
MCE truncation mutants were generated by PCR (Advantage KlenTag polymerase, CLONTECH) from pCR 2.1 MCE with a primer that introduces a BamHI site at the 5′ end and an antisense primer that inserts a stop codon and XhoI site at the 3′ end. The PCR products were cloned into pCR 2.1 (Invitrogen) and then subcloned into glutathione S-transferase (GST)-gene-fusion vector pGEX-4T-3 (Pharmacia). The pET28a-MCE (211–597) K294A mutant was constructed by site-specific mutagenesis (Quikchange Kit, Stratagene) utilizing pET-28a MCE (211–597) as template. Other MCE site-directed mutants were constructed by the same method from pGEX-4T-3 MCE (1–237) or pGEX-4T-3 MCE (211–597). Mutations were verified by DNA sequencing.
For expression of human capping enzyme (HCE) in insect cells, the BamHI-HindIII fragment from pCR2.1 HCE was inserted into polyhedrin promoter-based vector pBlueBacHis2A (Invitrogen). For subcellular localization studies of MCE (1–597), (1–237), and (211–597) in transfected mammalian cells, BamHI-BamHI fragments from pCR 2.1 MCE (1–597) and the other two corresponding constructs were inserted into myc-fusion vector pcB6+2myc, which contains two copies of the epitope sequence, EQKLISEEDL.
Expression and Purification of MCE Recombinant Proteins.
Escherichia coli BL21 (DE3) transformed with pET-28a MCE (211–597) K294A was grown at 37°C in Luria–Bertani (LB) medium containing kanamycin (50 μg/ml) and induced by addition of 0.4 mM isopropyl β-d-thiogalactoside (IPTG) after the culture reached A600 = 0.4. Cells were harvested after 3 hr and disrupted by sonication in lysis buffer [50 mM Tris, pH 8.5/10 mM 2-mercaptoethanol/1 mM phenylmethylsulfonyl fluoride (PMSF)]. Soluble proteins were incubated with Ni-NTA agarose (Qiagen) equilibrated with buffer A (20 mM Tris, pH 8.5/100 mM KCl/10 mM 2-mercaptoethanol/10% glycerol/20 mM imidazole). The agarose was washed with buffer A followed by buffer B (20 mM Tris, pH 8.5/1 M KCl/10 mM 2-mercaptoethanol/10% glycerol). Bound proteins were eluted with buffer C (20 mM Tris, pH 8.5/100 mM KCl/10 mM 2-mercaptoethanol/10% glycerol/100 mM imidazole).
E. coli BL21 (DE3) harboring pGEX-4T-3 MCE mutant constructs were grown at 30°C in LB medium containing ampicillin (100 μg/ml) and induced at A600 = 0.4 by the addition of 10 μM IPTG. Cells were harvested after 9–10 hr, and lysates were prepared by sonication in lysis/wash buffer (20 mM Tris, pH 7.9/0.1 M NaCl/0.2 mM EDTA/1 mM DTT/1 mM PMSF/0.2% Nonidet P-40). Soluble proteins were incubated in batches with glutathione-agarose (Sigma) equilibrated with lysis/wash buffer. The agarose then was washed extensively with lysis/wash buffer, and bound proteins were eluted with elution buffer (50 mM Tris, pH 8.0/10 mM reduced glutathione).
Expression and Purification of HCE Recombinant Proteins.
pBlueBacHis2A HCE was liposome-cotransfected with Bac-N-Blue DNA into Sf9 insect cells (Bac-N-Blue Transfection Kit, Invitrogen). Recombinant virus was detected on 5-bromo-4-chloro-3-indolyl β-d-galactoside plates and confirmed by PCR analysis. Sf9 cells infected with high-titer stock of the recombinant virus [multiplicity of infection (moi) = 7.5] were harvested after 2 days. Cell lysates were prepared by sonication in lysis buffer [50 mM Tris, pH 8.5/10 mM 2-mercaptoethanol/1 mM PMSF/1% Nonidet P-40/0.5 μg/ml aprotinin (Ap)/0.5 μg/ml leupeptin (Le)]. Soluble recombinant proteins were passed through a column of Ni-NTA agarose (Qiagen) and washed with buffer A plus Ap and Le and buffer B plus Ap and Le. Proteins were collected in buffer C plus Ap and Le.
All proteins were dialyzed against 20 mM Hepes, pH 7.5/1 mM EDTA/1 mM PMSF/5 mM DTT/20% glycerol/0.5% Nonidet P-40/100 mM KCl and then analyzed by SDS/PAGE with Coomassie blue staining. Protein concentrations were determined by Bio-Rad DC Protein Assay with BSA as standard.
RNA RTP Assay.
RTP substrates were synthesized as described (22) on BamHI-linearized pGEM1 (Promega) by T7 RNA polymerase (Promega) with 30 μCi [γ-32P]GTP (30 Ci/mmol, Dupont). Labeled RNA was incubated for 15 min at 37°C with 100 ng of the indicated proteins in 20 μl of 25 mM Tris, pH 7.5, containing 0.5 mM DTT. Reaction products were extracted with phenol/chloroform and analyzed on polyethyleneimine (PEI)-cellulose plates by TLC. The release of 32Pi was quantitated by PhosphorImager (Molecular Dynamics).
Modification of RTP Active Site.
The indicated proteins (500 ng) were modified as described (38) by incubation with 2 mM sodium iodoacetate (IOAc) or 2 mM iodo[2-14C]acetic acid (50 mCi/mmol, Amersham) in 20 μl of 50 mM 3,3-dimethyl glutarate/1 mM EDTA/0.15 mM NaCl for 30 min at 37°C. Products were analyzed for RTP activity or for radiolabeling by SDS/PAGE followed by autoradiography.
RNA Guanylyltransferase Assay.
The indicated proteins were incubated with 25 mM Tris, pH 7.5/5 mM MgCl2/0.5 mM DTT/0.1 μg inorganic pyrophosphatase (Boehringer Mannheim)/10 μCi [α-32P]GTP (3,000Ci/mmol, Amersham) for 15 min at 37°C. Guanylylation of proteins was assessed by SDS/PAGE followed by autoradiography.
Immunofluorescence Staining of MCE in Transfected Cells.
Mouse 3T3 cells were transfected with pcB6+2myc containing MCE (1–597), (1–237), or (211–597) sequences (SuperFect Transfection Reagent, Qiagen) and seeded into 8-cell chamber slides 24 hr before fixation and incubation for 2 hr with either anti-c-myc antibody (Oncogene Science, 1:300 dilution) or anti-MCE antibody (22). The secondary antibody, fluorescein isothiocyanate-conjugated goat anti-mouse IgG (Boehringer Mannheim, 1:100 dilution) or rhodamine-conjugated goat anti-rabbit IgG (Calbiochem, 1:100 dilution), was added for 1 hr before mounting the slides in aqua-polymount (Polyscience) and visualization by fluorescence.
Capping Enzyme Binding to Poly (U).
The indicated proteins were incubated with AGpoly (U) Type 6 (Pharmacia) in buffer D (150 mM Tris, pH 8.4/2 mM 2-mercaptoethanol/1 mM EDTA/10% glycerol/50 mM NaCl) for 1 hr at 4°C. The agarose was washed extensively with buffer D containing 0.2% Nonidet P-40. The bound proteins were analyzed by 10% SDS/PAGE, followed by silver staining or Western blot with anti-MCE antibody (22) or monoclonal anti-GST (Sigma) by Renaissance Chemiluminescence Reagent (NEN) and quantitated by Bio-Rad Imaging Densitometer.
RESULTS
The PTP Motif and Upstream Amino Acids Are Required for Mouse RTP Activity.
MCE contains at positions 124–134 a motif (I/V)HCXXGXXR(S/T)G conserved among PTPs. It includes the PTP active-site Cys, which also was shown to be essential for RNA 5′-triphosphatase activity of the N-terminal fragment of C. elegans capping enzyme (27) and, more recently, for other RTPs (23, 39, 40). We constructed and tested several truncation mutants of MCE to define the boundaries of its RTP domain. Constructs MCE (211–597) and (188–597) were inactive, while mutants MCE (1–237), (1–188), and (1–162), which contained the motif, had RTP levels similar to the full-length 597-aa protein (data not shown). Thus, the sequence (I/V)HCXXGXXR(S/T)G apparently is essential for mouse RTP activity.
C126 and D66, H125, R132, and T133 Are All Important for RTP Activity.
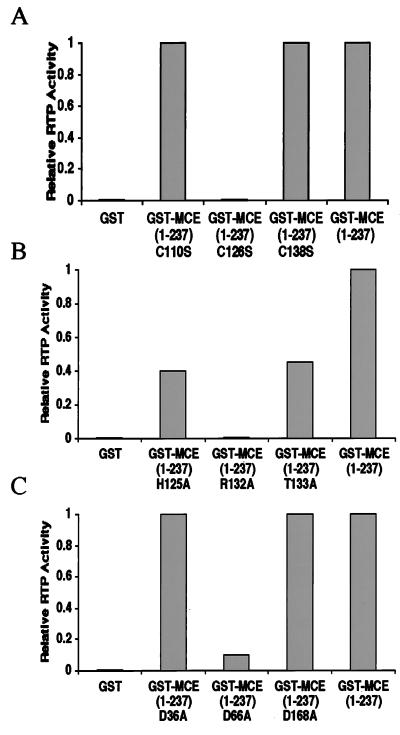
The presence of similar amino acid sequences in RTP and PTPs suggested that specific residues required in PTPs may also be necessary for RTP function. To test whether the motif C126 is essential for RTP activity, MCE (1–237) C126S was compared with two mutant proteins with substitutions in motif-proximal cysteines at positions 110 and 138. MCE (1–237) C110S and MCE (1–237) C138S were fully active, while MCE (1–237) C126S was completely devoid of RTP activity (Fig. 1A). These results demonstrate that the motif Cys likely is involved in the formation of a Cys-P intermediate (as in PTPs) and that the loss of activity in the C126S mutant probably is not due to the absence of an essential disulfide.
Figure 1.
C126, R132, H125, T133, and D66 are important for mouse RTP activity. γ-32P-labeled RNA was incubated with 100 ng of the indicated purified proteins as described in Materials and Methods. Released 32Pi was quantitated by PhosphorImager. Values are expressed relative to 1.0 for GST-MCE (1–237). Variation among several experiments was ±0.1.
Because the motif Arg, His, and Thr residues are also important for PTP function, we assayed MCE (1–237) point mutants R132A, H125A, and T133A for RTP activity (Fig. 1B). No detectable 32Pi was released by the R132A mutant, and the activity of the H125A and T133A mutants was decreased by 55–60% compared with MCE (1–237). PTPs also require an Asp located outside the conserved motif to act as a general acid and facilitate release of Pi (30–32, 41). Alignment of human, mouse, and C. elegans capping enzymes with human 1B and baculovirus BVP PTPs suggested that D36, D66, and D168 in MCE corresponded to conserved residues. Ala substitution at positions 36 or 168 was without effect, but the RTP activity of the D66A mutant was decreased by more than 90% (Fig. 1C). These single-site mutant analyses indicate that the amino acids essential for mouse RTP include corresponding residues required for catalytic activity by PTPs.
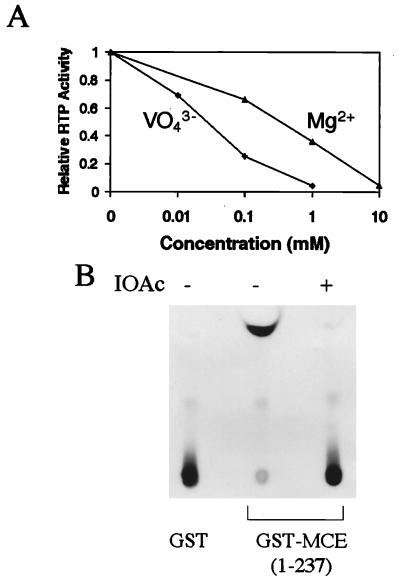
VO43−, Mg2+, and Iodoacetate Inhibit Mouse RTP.
Vanadate covalently binds to the active-site Cys in Yersinia PTP and acts as a transition-state mimic and inhibitor (30, 31). Vanadate also inhibited mouse RTP in a concentration-dependent fashion, and at 1 mM Na3VO4 the activity was decreased by more than 95% (Fig. 2A). PTPs were shown previously to be independent of metal ions (31), and mouse RTP not only did not require Mg2+ but was inhibited by the divalent cation (Fig. 2A).
Figure 2.
VO43−, Mg2+, and IOAc inhibit mouse RTP activity. Purified GST-MCE (1–237) was assayed for RTP activity as in Fig. 1 in the presence of the indicated concentrations of Na3VO4 or MgCl2 (A) or IOAc (B).
IOAc is an irreversible sulfhydryl-directed enzyme inhibitor that can completely inactivate PTPs by specifically modifying the conserved motif active-site Cys (38). To test whether the C126 in mammalian RTP is similarly sensitive, purified MCE (1–237) was incubated with sodium IOAc. This treatment reduced enzyme activity essentially completely (Fig. 2B). In addition, iodo[2-14C]acetic acid radiolabeled MCE (1–237) but not the C126S mutant (data not shown), suggesting that among the total of 7 cysteines in MCE (1–237), only position 126 is readily available for carboxymethylation. These results provide further evidence for use of a similar (or identical) enzymatic mechanism by RTP and PTPs.
Full-Length Capping Enzyme and the C-Terminal Guanylyltransferase Fragments Are Localized in Nuclei.
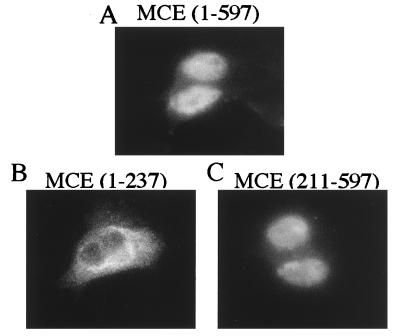
To investigate the subcellular localization of capping enzyme and its domains, 3T3 cells were transfected with the myc-fusion vector encoding MCE (1–597), (1–237), or (211–597). Staining for myc-fusion proteins demonstrated that full-length capping enzyme and the C-terminal guanylyltransferase fragment were mainly nuclear, consistent with their binding to pol II (22). By contrast, the N-terminal RTP fragment was mostly cytoplasmic (Fig. 3). The results suggest that C-terminal guanylyltransferase and not the N-terminal RTP contains sequences required for nuclear localization of mammalian capping enzyme.
Figure 3.
Full-length capping enzyme and C-terminal guanylyltransferase fragment are localized in nuclei. 3T3 cells transfected with pcB6+2myc containing MCE (1–597) (A), (1–237) (B), or (211–597) (C) were analyzed by immunofluorescence staining as described in Materials and Methods.
Full-Length Capping Enzyme and the C-Terminal Guanylyltransferase Domain Bind Poly (U) Beads.
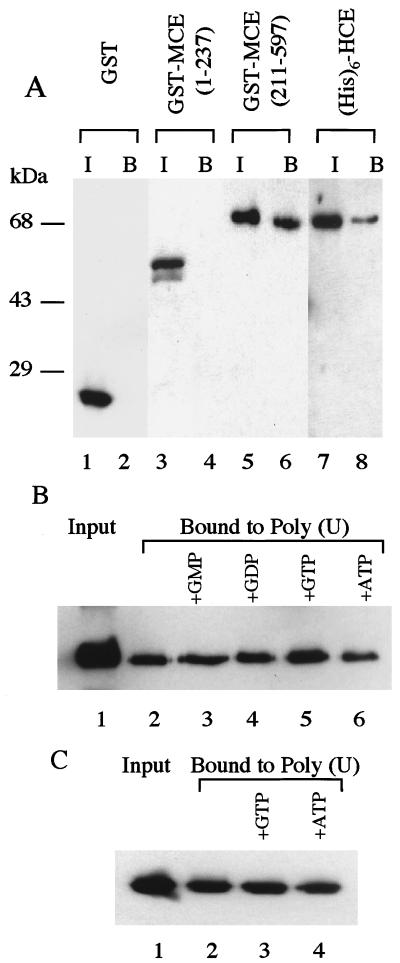
Previous studies implied that capping enzymes recognize primary transcripts by selective association with RNA pol IIo (22, 33, 34). To investigate whether capping enzyme also interacts with RNA directly, purified fusion proteins (His)6-HCE, GST-MCE (1–237), and GST-MCE (211–597) were incubated with poly (U) beads, and bound proteins were analyzed by SDS/PAGE followed by silver staining (data not shown) or Western blot. As shown in Fig. 4A, control GST (lane 2) and GST-MCE (1–237) (lane 4) did not bind to poly (U), while GST MCE (211–597) (lane 6) and (His)6-HCE (lane 8) bound to the ribopolymer. These results indicate that the C-terminal guanylyltransferase, but not the N-terminal RTP domain of capping enzyme, can interact directly with RNA.
Figure 4.
Full-length capping enzyme and C-terminal guanylyltransferase fragment bind poly (U). (A) Purified GST, GST-MCE (1–237), GST-MCE (211–597), and (His)6-HCE were incubated with poly (U) beads as described in Materials and Methods. Bound proteins (lanes 2, 4, 6, and 8) were analyzed by SDS/PAGE and Western blot in comparison with the input (lanes 1, 3, 5, and 7). Immunoblots were developed by anti-GST antibody (lanes 1–6) or anti-MCE antibody (lanes 7 and 8). (B) Purified GST-MCE (211–597) (lane 1) was incubated with poly (U) beads (lane 2) in the presence of 30 μM each of GMP (lane 3), GDP (lane 4), GTP (lane 5), or ATP (lane 6). Bound proteins were analyzed by SDS/PAGE and Western blot analysis with anti-MCE antibody. (C) Purified (His)6-MCE (211–597) K294A (lane 1) was incubated with poly (U) beads (lane 2) and 30 μM GTP (lane 3) or ATP (lane 4) followed by analysis as in B.
The C-terminal guanylyltransferase domain of MCE binds poly (U), and the presence of GTP reproducibly increased the binding of GST-MCE (211–597) by ≈1.4-fold; GMP, GDP, and ATP had little or no effect (Fig. 4B). Binding assays also were done with mutant protein (His)6-MCE (211–597) K294A in which the guanylyltransferase active-site lysine 294 was replaced by Ala. Although the inactive mutant protein bound poly (U) as well as wild-type enzyme, the addition of GTP had no effect (Fig. 4C), consistent with enhancement of RNA binding by guanylylation.
Involvement of Residues R315, R530, K533, and N537 in Both Poly (U) Binding and Guanylyltransferase Activity.
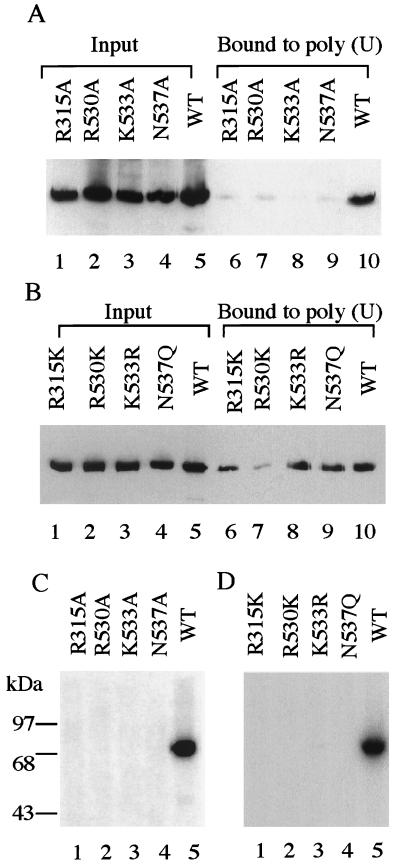
In an effort to identify amino acids involved in RNA binding, we compared the sequences of mouse and Chlorella virus PBCV-1 capping enzymes. By fitting the amino acid sequence of the mouse enzyme into the three-dimensional structure of the Chlorella virus protein using the homology modeling program modeller (42), MCE residues R315, R530, K533, and N537 were positioned at the GTP-binding pocket entrance. Consistent with this location and involvement in RNA binding, Ala substitution at each of these four positions eliminated poly (U) binding almost completely (Fig. 5A). Conservative single-site substitutions were made in MCE (211–597), including R315K, R530K, K533R, and N537Q. All but R530K retained 60% or more of the RNA binding (Fig. 5B). These data suggest that direct association between the guanylyltransferase domain and poly (U) depend on the positive charge or, in the case of N537, the polar hydrophilic character at these positions.
Figure 5.
R315, R530, K533, and N537 are involved in both poly (U) binding and guanylylation. (A and B) Purified GST-MCE (211–597) (lane 5) and the indicated mutants (lanes 1–4) were incubated with poly (U) beads, and bound proteins (lanes 6–10) were analyzed in comparison with the input as in Fig. 4A. (C and D) Purified GST-MCE (211–597) and the indicated mutants were assayed for guanylyltransferase activity by incubation with [α-32P]GTP followed by SDS/PAGE and autoradiography, as described in Materials and Methods.
Because molecular modeling suggested that R315, R530, K533, and N537 are located at the mouth of the GTP-binding pocket of mouse guanylyltransferase, we examined whether these residues are also involved in guanylyltransferase activity. Purified MCE (211–597) mutant proteins were incubated with [α-32P]GTP and analyzed by SDS/PAGE. Ala substitutions or conservative replacements at each of these four positions eliminated guanylyltransferase activity (Fig. 5 C and D). Thus, although charge and hydrophilicity at positions R315, R530, K533, and N537 are important for poly (U) binding, additional characteristics also play a role in guanylyltransferase activity.
DISCUSSION
Cotranscriptional 5′ capping of pol II transcripts is crucial for eukaryotic gene expression. Mammalian and C. elegans capping enzymes are bifunctional proteins in which the N-terminal RTP domain contains the (I/V)HCXXGXXR(S/T)G motif characteristic of PTPs (31, 41). In addition to the motif Cys shown initially to be required for RTP activity in C. elegans (27), we identified D66, H125, R132, and T133 as important for mouse RTP activity and showed that vanadate and IOAc are inhibitory, as in PTPs. These results are consistent with mammalian RTP as well as PTPs using the PTP mechanism for phosphoryl group removal from RNA and proteins, respectively. In this mechanism (31, 32, 41, 43) the conserved C126 of mouse RTP would make a nucleophilic attack on the γ-phosphate of the 5′ end of pol II primary transcripts to generate a Cys-P intermediate. The phosphate subsequently is transferred to a water molecule activated by D66, which also enhances intermediate formation by acting as a general acid. R132 in the conserved motif may contribute to the stabilization of the RTP intermediate as in PTP transition-state Cys-P intermediates.
Previous studies have shown that capping enzymes selectively associate with the hyperphosphorylated CTD of RNA pol II (33, 34). Nuclear localization of the full-length and C-terminal but not the N-terminal domain of MCE agrees with the findings that the C-terminal guanylyltransferase domain of MCE was sufficient for pol II binding (22) and with the absence of detectable interaction between the N-terminal RTP domain and pol II. These data imply that capping enzymes are recruited to nascent transcripts by interaction with pol II. The present study demonstrates that MCE also can interact directly with RNA and that the C-terminal guanylyltransferase domain, rather than the N-terminal RTP, mediates this interaction. Selective capping of pol II products may be a two-step process involving access of guanylyltransferase to nascent transcripts, indirectly by pol II CTD binding and directly by RNA binding. Thus, in metazoans the N-terminal triphosphatase is targeted to RNA 5′ ends by its physical linkage to the guanylyltransferase domain in a single, bifunctional polypeptide. Recently, it was observed that full-length HCE complements mutant S. cerevisiae lacking the essential RTP activity, while C-terminal truncation mutants that retained triphosphatase activity were unable to substitute for the deleted yeast RTP (26). A possible explanation for these results is that RTP relies on both RNA- and pol II-binding sites in the guanylyltransferase to recognize the newly initiated pol II transcripts. The HCE C-terminal truncation mutants that were tested for complementation were missing the region involved in binding and, probably as a consequence, were unable to access the RNA substrates for γ-phosphate hydrolysis. Alternatively, residues 285-PPPPPKR-292 in HCE may correspond to a nuclear localization signal, their deletion in the C-terminal truncation mutants resulting in loss of complementation because of failure of the truncated protein to localize to nuclei.
Binding of guanylyltransferase to poly (U) was increased reproducibly by GTP, and guanylylation of capping enzyme may enhance RNA binding. Ala substitution analyses showed that RNA binding requires at least R315, R530, K533, and N537, and they are also necessary for guanylyltransferase activity. Interestingly, conservative replacements (R315K, R530K, K533R, and N537Q) eliminated guanylyltransferase activity but not poly (U) binding. Previous studies of the x-ray crystal structure of Chlorella virus PBCV-1 capping enzyme revealed that capping enzyme has two different conformations, open and closed (37). On the basis of these and other results we suggest that MCE R315, R530, K533, and N537 may be part of the surface that interacts with RNA and with GTP. After addition of GMP to form the enzyme intermediate, the positions of these four residues relative to RNA and GMP may be shifted because of conformational changes in the guanylyltransferase domain. Loss of interaction with the GMP moiety of these (and possibly other) residues leads to stronger binding to RNA and GMP transfer to nascent transcripts.
Acknowledgments
We are grateful to Jae-Kyoung Hong, Dr. Kalyan Das, and Dr. Edward Arnold for providing a molecular model of MCE based on the x-ray structure of PBCV-1 capping enzyme. We thank Hao Chen for helpful advice and discussions. We also thank Helen Gu for purified (His)6-MCE (211–597) K294A protein, Dr. Cory Abate-Shen for pcB6+2myc vector, and Yujie Zhao and Gang Li for assistance in the immunofluorescence studies. We are indebted to Drs. Arnold, Céline Gélinas, and Arnold B. Rabson for critical reading of the manuscript.
ABBREVIATIONS
- Ap
aprotinin
- GST
glutathione S-transferase
- HCE
human capping enzyme
- IOAc
iodoacetate
- Le
leupeptin
- MCE
mouse capping enzyme
- PEI
polyethyleneimine
- PTP
protein tyrosine phosphatase
- RTP
RNA 5′-triphosphatase
- pol IIo
RNA polymerase II
References
- 1. Shatkin A J. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 2.Konarska M M, Padgett R A, Sharp P A. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 3.Krainer A R, Maniatis T, Ruskin B, Green M R. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 4.Shuman S. Prog Nucleic Acids Res Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- 5.Flaherty S M, Fortes P, Izaurralde E, Mattaj I W, Gilmartin G M. Proc Natl Acad Sci USA. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuichi Y, LaFiandra A, Shatkin A J. Nature (London) 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 7.Shimotohno K, Kodama Y, Hashimoto J, Miura K I. Proc Natl Acad Sci USA. 1977;74:2734–2738. doi: 10.1073/pnas.74.7.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo H, Huang H, Donahue T F. Mol Cell Biol. 1998;18:665–675. doi: 10.1128/mcb.18.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwer B, Mao X, Shuman S. Nucleic Acids Res. 1998;26:2050–2057. doi: 10.1093/nar/26.9.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamm J, Mattaj I W. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 11.Görlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey R A, Mattaj I W, Izaurralde E. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 12.Shatkin A J. Cell. 1985;40:223–224. doi: 10.1016/0092-8674(85)90132-1. [DOI] [PubMed] [Google Scholar]
- 13.Sonenberg N. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 245–269. [Google Scholar]
- 14.Morley S J, Curtis P S, Pain V M. RNA. 1997;3:1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 15.Moss B. In: Fields Virology. 3rd Ed. Fields B N, Knipe D, Howley P M, editors. Philadelphia: Lippincott; 1996. pp. 2637–2671. [Google Scholar]
- 16.Furuichi Y, Muthukrishnan S, Tomasz J, Shatkin A J. J Biol Chem. 1976;251:5043–5053. [PubMed] [Google Scholar]
- 17.Moss B, Gershowitz A, Wei C, Boone R. Virology. 1976;72:341–351. doi: 10.1016/0042-6822(76)90163-x. [DOI] [PubMed] [Google Scholar]
- 18.Shibagaki Y, Itoh N, Yamada H, Nagata S, Mizumoto K. J Biol Chem. 1992;267:9521–9528. [PubMed] [Google Scholar]
- 19.Shuman S, Liu Y, Schwer B. Proc Natl Acad Sci USA. 1994;91:12046–12050. doi: 10.1073/pnas.91.25.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada-Okabe T, Shimmi O, Doi R, Mizumoto K, Arisawa M, Yamada-Okabe H. Microbiology. 1996;142:2515–2523. doi: 10.1099/00221287-142-9-2515. [DOI] [PubMed] [Google Scholar]
- 21.Tsukamoto T, Shibagaki Y, Imajoh-Ohmi S, Murakoshi T, Suzuki M, Nakamura A, Gotoh H, Mizumoto K. Biochem Biophys Res Commun. 1997;239:116–122. doi: 10.1006/bbrc.1997.7439. [DOI] [PubMed] [Google Scholar]
- 22.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin A J. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho C K, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. J Biol Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 24.Pillutla R C, Yue Z, Maldonado E, Shatkin A J. J Biol Chem. 1998;273:21443–21446. doi: 10.1074/jbc.273.34.21443. [DOI] [PubMed] [Google Scholar]
- 25.Tsukamoto T, Shibagaki Y, Murakoshi T, Suzuki M, Nakamura A, Gotoh H, Mizumoto K. Biochem Biophys Res Commun. 1998;243:101–108. doi: 10.1006/bbrc.1997.8038. [DOI] [PubMed] [Google Scholar]
- 26.Yamada-Okabe T, Doi R, Shimmi O, Arisawa M, Yamada-Okabe H. Nucleic Acids Res. 1998;26:1700–1706. doi: 10.1093/nar/26.7.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takagi T, Moore C R, Diehn F, Buratowski S. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 28.Wang S P, Deng L, Ho C K, Shuman S. Proc Natl Acad Sci USA. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fresco L D, Buratowski S. Proc Natl Acad Sci USA. 1994;91:6624–6628. doi: 10.1073/pnas.91.14.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denu J M, Lohse D L, Vijayalakshmi J, Saper M A, Dixon J. Proc Natl Acad Sci USA. 1996;93:2493–2498. doi: 10.1073/pnas.93.6.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fauman E B, Saper M A. Trends Biochem Sci. 1996;21:413–417. doi: 10.1016/s0968-0004(96)10059-1. [DOI] [PubMed] [Google Scholar]
- 32.Bell S D, Denu J M, Dixon J E, Ellington A D. J Biol Chem. 1998;273:14309–14314. doi: 10.1074/jbc.273.23.14309. [DOI] [PubMed] [Google Scholar]
- 33.Cho E, Takagi T, Moore C R, Buratowski S. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D L. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagler J, Shuman S. Science. 1992;255:983–986. doi: 10.1126/science.1546295. [DOI] [PubMed] [Google Scholar]
- 36.Luo Y, Shuman S. J Biol Chem. 1993;268:21253–21262. [PubMed] [Google Scholar]
- 37.Håkansson K, Doherty A J, Shuman S, Wigley D B. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 38.Pot D A, Dixon J E. J Biol Chem. 1992;267:140–143. [PubMed] [Google Scholar]
- 39.Takagi T, Taylor G S, Kusakabe T, Charbonneau H, Buratowski S. Proc Natl Acad Sci USA. 1998;95:9808–9812. doi: 10.1073/pnas.95.17.9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gross C H, Shuman S. J Virol. 1998;72:7057–7063. doi: 10.1128/jvi.72.9.7057-7063.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tonks N K, Neel B G. Cell. 1996;87:365–368. doi: 10.1016/s0092-8674(00)81357-4. [DOI] [PubMed] [Google Scholar]
- 42.Sali A, Blundell T L. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 43.Denu J M, Stuckey J A, Saper M A, Dixon J E. Cell. 1996;87:361–364. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]