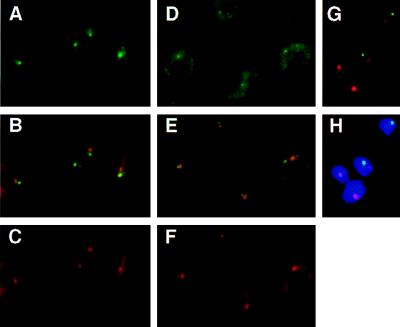
Figure 3.
(A–C) Subnuclear localization of the nuclear transcripts derived from the active VSG ES. T. brucei variant 221a cells hybridized with probe mix 4 (see Fig. 1A) and the ribosomal probe R2. Cells were treated with DNase before hybridization. (A) Detection of probe mix 4 (Fig. 1A and Materials and Methods) with FITC-conjugated antibodies. The single intense spot, in the nucleus, shows the localization of the nascent transcripts derived from the active site. The more diffuse staining in the cytoplasm corresponds to ES mRNAs. (B) Simultaneous detection of the active ES RNA (green) and the nucleolus (red). Overlap between the two signals appears in yellow. (C) Detection of the ribosomal probe R2 (Materials and Methods), with Texas Red-conjugated antibodies. The circle in the nucleus shows the signal derived from the nuclear rRNA, forming the nucleolus (together with the rDNA and the processing machinery). The cytoplasmic signal comes from the rRNA in ribosomes. (D–H) Subnuclear localization of marker genes integrated in the active VSG ES in the 3174 transformant of T. brucei variant 221a (ref. 36; Fig. 1B). (D) Detection of probe neo+hyg (active VSG ES) with FITC-conjugated antibodies. The probe hybridized to the marker genes appears as a small fluorescent signal in the nucleus; a cytoplasmic signal attributable to mRNA is also visible. (E) Simultaneous detection of the active ES (green) and the rDNA in the nucleolus (red). The overlap of signals appears as yellow (or slightly orange). (F) Detection of the signal derived from the ribosomal probe R2. (G and H) Simultaneous detection of the active ES (green) and the rDNA (red) in RNase-treated cells; H also shows the DAPI-stained nucleus).