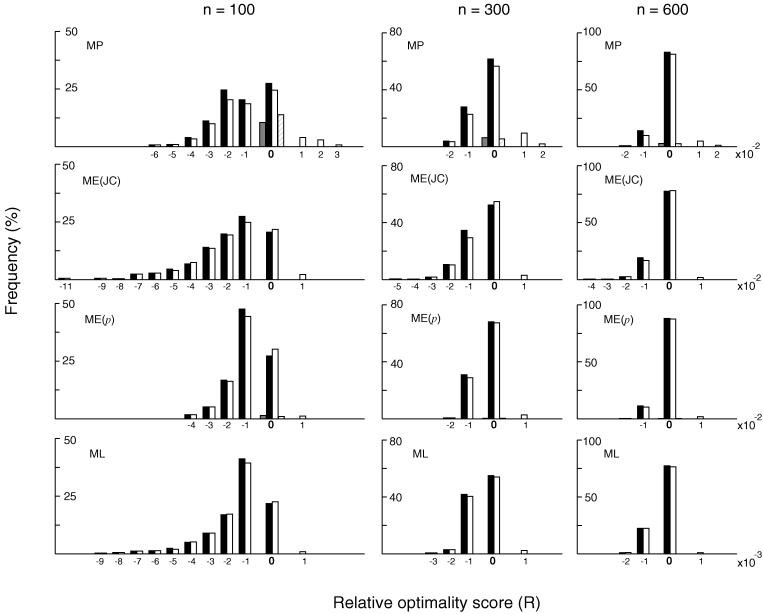
Figure 2.
Distributions of relative optimality scores (R) of the MP, ME, and ML trees inferred by the exhaustive search (solid bars) and the single-tree search (open bars) algorithms. These results were obtained from 500 replications of computer simulation following model tree A in Fig. 1 with dmax = 1.0. n represents the number of nucleotides used. ME(JC) and ME(p) refer to ME trees with the Jukes–Cantor distance and the p-distance, respectively. R values for MP and ME (or NJ) trees are multiplied by c = 100, whereas those for ML trees are multiplied by c = 1,000. cR = 0 represents the case where the correct topology was obtained. Except for cR = 0, cR = x represents the cR values in the range of x − 1 < cR ≤ x for a positive integer x and in the range of x ≤ cR < x + 1 for a negative integer x. Thus, cR = 1 represents the cR values between 0 and 1 excluding cR = 0, cR = 2 represents the cR values between 1 and 2 excluding cR = 1, and cR = −1 represents the cR values between −1 and 0 excluding 0. We used c = 1,000 for ML trees, because the scale of R for ML trees was much finer than that for MP and ME trees.