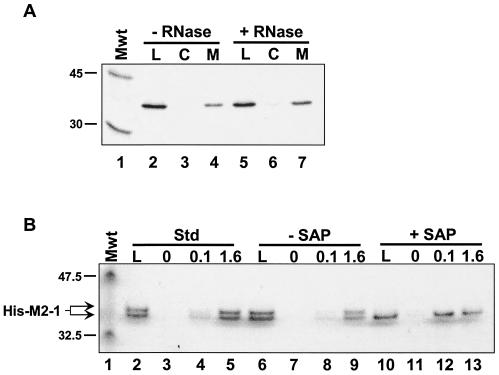
FIG. 3.
The M2-1-P interaction is not mediated by RNA or M2-1 phosphorylation. (A) Immobilized His-M2-1 and IVT P proteins were both incubated either in the absence (− RNase) or presence (+ RNase) of a mixture of RNase A and RNase T1. Affinity chromatography was performed as described in the legend to Fig. 1. Lanes: L, 1/20 of the material loaded on each column; C, high-salt elutions from the control columns; M, salt elutions from the M2-1 columns. The positions of the 45-kDa (ovalbumin) and 30-kDa (carbonic anhydrase) from 14C-labeled rainbow molecular mass markers (Mwt) (Amersham) are indicated to the left of the gel. (B) His-M2-1 was diluted in affinity column buffer (Std, standard) (lanes 2 to 5) or 1× SAP (Promega) buffer (lanes 6 to 13). In the latter case, the protein was incubated either in the absence or presence of SAP, as indicated, prior to affinity chromatography on columns containing 0, 0.1, or 1.6 mg of immobilized GST-P per ml, as indicated above the gel. High-salt eluates were analyzed on a 4 to 12% NuPAGE (Invitrogen) gel run with 1× MES buffer, as suggested by the manufacturer. The gel was silver stained using the SilverSNAP kit (Pierce). The positions of the 47.5-kDa (aldolase) and 32.5-kDa (triose phosphate isomerase) proteins, which are prestained molecular mass markers (Mwt) (New England BioLabs), and the position of M2-1 are indicated to the left of the gel.