Figure 6.
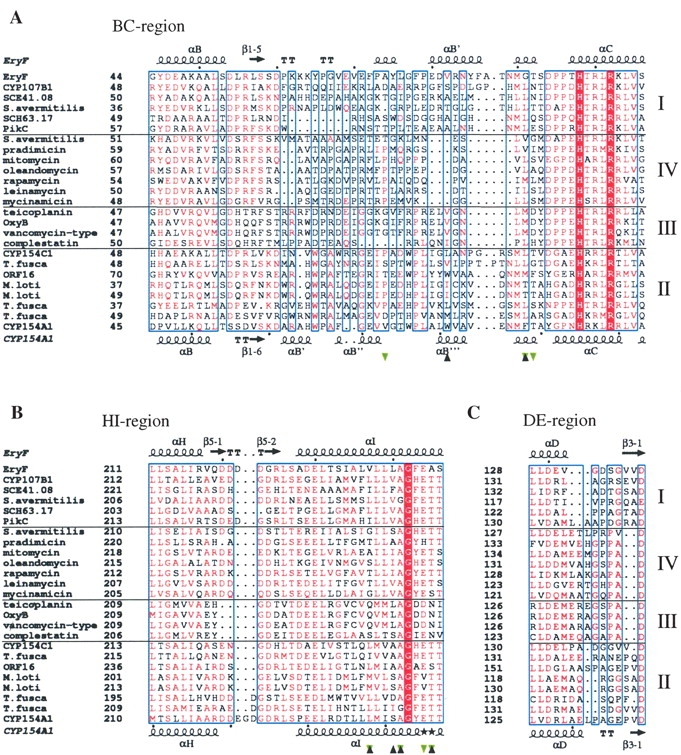
Fragments of CYP sequence alignment. (A) BC region, (B) HI region, and (C) DE region. The alignment was performed using MULTALIN (Corpet 1988; Gouet et al. 1999) with a similarity global score of 0.3 and a differentiation score of 0.3. Residue colors are according to the following rules: red box/white character, strict identity; red character, similarity in the group; blue frames, similarity across groups. Groups are defined based on similarity scores calculated using the algorithm implemented in the program. Branches of sequences are separated by solid horizontal lines and numbered on the right. Sequences are labeled at the left with the protein or gene name if available, and otherwise according to the biosynthetic pathway or organism of origin if anything else is unknown. Residues interacting with 4-phenylimidazole in CYP154A1 are labeled with black triangles; those interacting with 6-deoxyerythronolide B in Eryf are labeled with green triangles. Secondary structure elements are assigned based on 1OXA for Eryf and 1ODO for CYP154A1. Black stars in place of secondary structure element symbols label residues with alternate conformations. Black dots point at every tenth residue in the CYP154A1 and EryF sequences. The image was generated using the program ESPript (Gouet et al. 1999) The full sequence alignment is represented in the Supplemental Material.
