Figure 1.
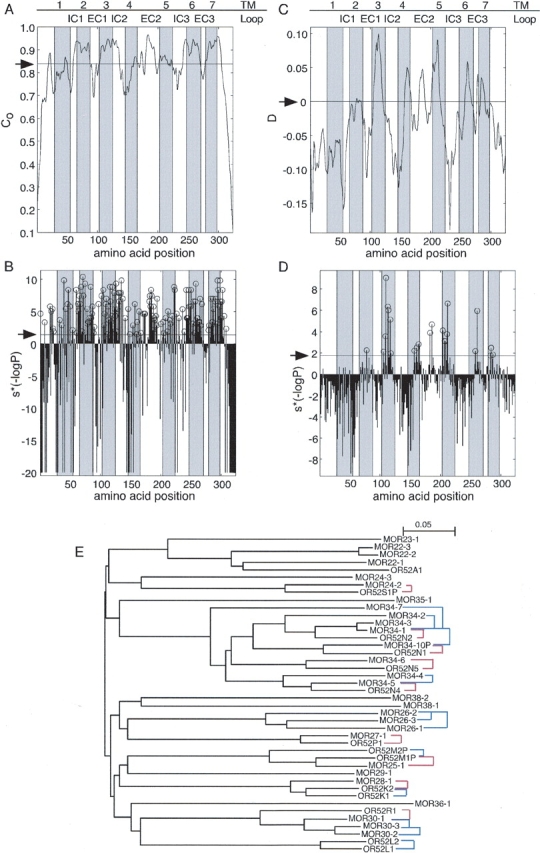
(A) Positional conservation within orthologous OR pairs computed along the multiple sequence alignment of 218 such pairs using equation 1. (B) The significance (P) of the positional conservation computed along the OR multiple sequence alignment. In the profile plotted, S*(−logP) is shown. S indicates whether the observed positional conservation is more (S = 1) or less (S = −1) than that expected by chance. Positions that are significantly conserved are marked with open circles. (C) The difference between the positional conservation within 218 orthologous OR pairs (Co) and that within 518 paralogous OR pairs (Cp), D, computed along the multiple sequence alignment. (D) The significance (P) of the difference D computed along the OR multiple sequence alignment. In the profile plotted, S*(−logP) is shown. S differentiates between positions for which D > 0 (S = 1) from positions for which D < 0 (S = −1). Positions that are significantly more conserved within orthologous pairs than within paralogous pairs are marked with open circles. The positions of TM segments, as inferred from rhodopsin, are shown as shaded areas. In A and C the arrow indicates the expectation value; in B and D it indicates the cutoff dictated by an FDR of 0.05. The original profiles in A and C were smoothed using the “hamming” function of the MATLAB/Math Works Inc. package with a window size = 7. (E) The phylogenetic relationships captured by the ortholog and paralogs sets. A neighbor-joining tree (Saitou and Nei 1987) is shown for selected ORs. Distances within the tree correspond to divergence between the receptors. Names of human ORs begin with OR, whereas those of mouse begin with MOR. Red lines indicate pairs from the ortholog set; blue lines indicate pairs from the paralog set. As can be seen, in some cases a receptor has more than one ortholog according to the tree. In such cases our ortholog selection criteria chose the ortholog with the highest sequence identity (least divergence). Thus, the selected pair was the one most likely to contain ORs that share similar odorant specificity.
