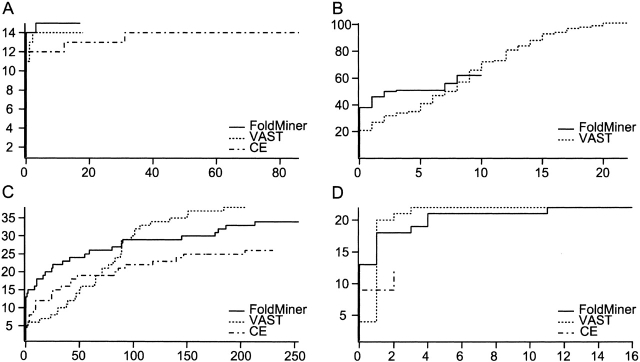
Figure 4.
Receiver operating characteristic (ROC) curves compare the performance of FoldMiner, VAST, and CE. Four representative ROC curves reveal trends observed in the full set of curves determined for all queries in seven different SCOP folds. VAST alignments are ranked by alignment score; minor changes occur if they are instead ranked by the number of aligned residues as described in the text. (A) The ROC curve for the globin query d1ew6a_ shows comparable performance of the three algorithms, although FoldMiner identifies one more member of the phycocyanin superfamily of the globin fold than do VAST and CE. (B) FoldMiner identifies more structural neighbors of the immunoglobulin query d1qfoa_ before it finds its first false positive than do the other algorithms, but finds fewer true positives overall than does VAST. CE returned no results with Z scores >3.7 in this particular case. (C) All three algorithms find a large number of false positives when the flavodoxin structure d5nul__ is used as the query, as the general motif of helices packing against both sides of a small β sheet is observed in many different folds. (D) FoldMiner and VAST perform equally well overall when the β-grasp structure d1vcba_ is used as the query, but FoldMiner more accurately ranks true positives ahead of false positives in early regions of the curve than does VAST.