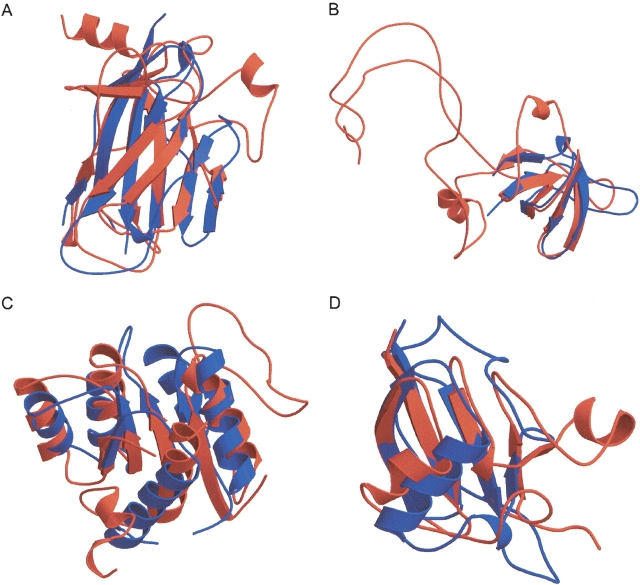
Figure 5.
LOCK 2 alignments frequently reveal structural similarities that cross SCOP fold and superfamily boundaries. (A) The immunoglobulin query d1neu__ (blue) aligns well to many Greek keys outside of the SCOP immunoglobulin fold; an alignment to d1ycsa_ (red) is shown. (B) An alignment of SCOP domain d1ckaa_ (blue), an SH3-domain, to d1d7qa_ (red), an OB-fold domain, reveals structural similarities between the two domains. Similarly, other SCOP folds that contain barrels frequently align well to domains in the SH3-like barrel fold. (C) Many members of the SCOP flavodoxin-like fold show strong structural similarity to the NAD(P)-binding Rossman fold. Here, a flavodoxin (d3chy__, shown in blue) is aligned to d1dih_1 (red). (D) An alignment of a human protein from SCOP’s ubiquitin-like superfamily (d1vcba_, shown in blue) to a 2E–2S ferredoxin from a cyanobacterium (d1czpa_, shown in red) reveals structural similarities between two superfamilies of SCOP’s β-Grasp (ubiquitin-like) fold. This figure was produced by Molscript and rendered by Raster3D (Kraulis 1991; Esnouf 1997; Merritt and Bacon 1997).