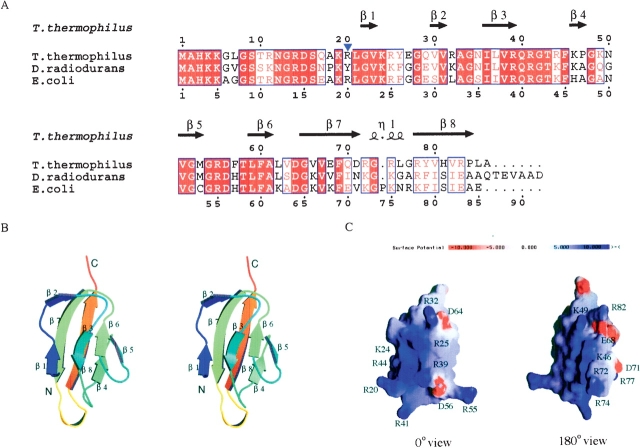
Figure 1.
(A) Alignment of the ribosomal protein L27 sequences from T. thermophilus HB8, D. radiodurans, and E. coli. The multiple alignment was achieved with CLUSTAL X (Thompson et al. 1997). The secondary structural elements, determined on the basis of the X-ray structure of T. thermophilus protein L27, are indicated with arrows for β-strands and a coil for a 310 helix at the top of the alignment. The identical amino acid residues are shown in white letters highlighted in red, whereas the similar residues are shown in red letters. (B) Stereo structure of the T. thermophilus protein L27. The stereo views were prepared by the programs MOLSCRIPT (Kraulis et al. 1991) and RASTER3D (Merritt and Bacon 1997). (C) Electrostatic potential surface of the T. thermophilus ribosomal protein L27. The molecular surface presentation was generated by GRASP (Nicholls et al. 1991).