Figure 1.
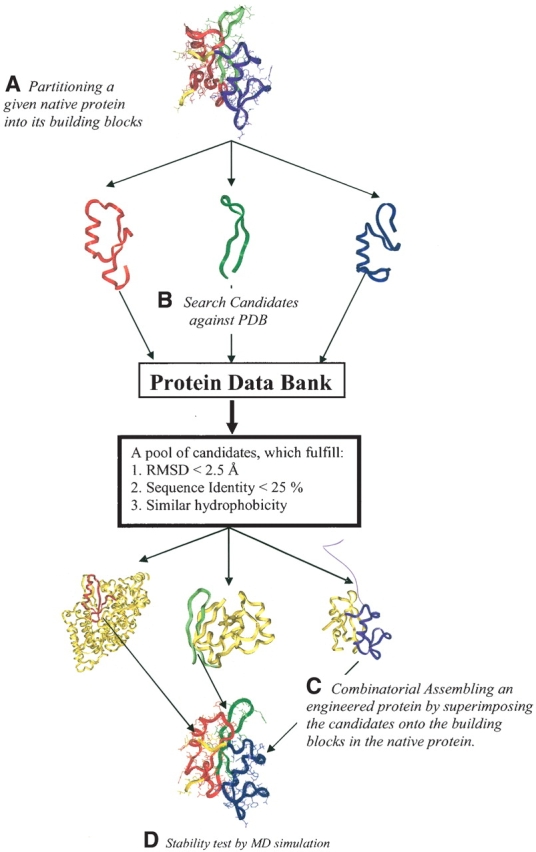
Graphic outline of the computational protein engineering algorithm. (A) A given native protein 3D structure is partitioned into building blocks using a computational cutting algorithm and a fragment length independent scoring function (Tsai et al. 2000). (B) Using the structure and sequence of building blocks from step A, candidate substitution fragments are searched against the PDB. The goal is a Cα-RMSD smaller than 2.5 Å and sequence identity lower than 25%. At the same time, candidate building blocks should have a similar hydrophobic/hydrophilic pattern. (C) By superimposing the selected substitution fragments onto the native protein, the new engineered protein is constructed by combinatorial assembly. (D) Finally, the stability of the engineered proteins are examined by explicit water MD simulations. This algorithm ensures the engineered protein has the similar fold as its parent protein, but with low sequence identity.
