Table 4.
ATP-dependent kinetic parameters for wild-type and mutant CPS
| Glutamine-dependent ATPase |
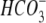 -dependent ATPase -dependent ATPase |
CP-dependent ATP synthesis | |||||||
| Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) | |
| WT | 0.029 ± 0.002 | 4.04 ± 0.1 | 137.5 | 0.005 ± 3.10 • 10−4 | 0.09 ± 2.9 • 10−3 | 17.9 | 0.006 ± 0.001 | 0.36 ± 0.01 | 65.3 |
| A144Q | 0.285 ± 0.056 | 3.94 ± 0.51 | 13.9 | 0.034 ± 0.007 | 0.10 ± 0.01 | 2.9 | 0.005 ± 3.9 • 10−5 | 0.28 ± 8.0 • 10−4 | 56.4 |
| D207A | 0.015 ± 0.002 | 1.51 ± 0.11 | 100.9 | 0.006 ± 0.001 | 0.11 ± 0.01 | 18.9 | 0.006 ± 0.001 | 0.26 ± 0.02 | 44.6 |
| S209A | 0.031 ± 0.004 | 2.43 ± 0.13 | 78.0 | 0.016 ± 0.001 | 0.35 ± 0.01 | 22.7 | 0.011 ± 0.001 | 0.59 ± 0.01 | 51.4 |
| ADS | 0.571 ± 0.083 | 0.66 ± 0.04 | 1.1 | 0.319 ± 0.012 | 0.33 ± 0.01 | 1.0 | 0.008 ± 0.001 | 0.28 ± 0.01 | 36.3 |
| D207N | 0.020 ± 0.018 | 1.65 ± 0.02 | 84.2 | 0.009 ± 0.003 | 0.06 ± 0.01 | 7.2 | 0.004 ± 0.002 | 0.14 ± 0.01 | 36.6 |
| A144Q/D207A | 0.154 ± 0.001 | 1.70 ± 0.08 | 11.0 | 0.021 ± 0.002 | 0.12 ± 0.01 | 5.8 | 0.011 ± 4.0 • 10−4 | 0.37 ± 0.04 | 33.9 |
| I211S | 0.072 ± 0.009 | 4.77 ± 0.26 | 66.4 | 0.020 ± 0.002 | 0.29 ± 0.01 | 14.4 | 0.011 ± 0.002 | 0.48 ± 0.02 | 44.0 |
| P690Q | 0.383 ± 0.061 | 9.01 ± 0.79 | 23.5 | 0.009 ± 0.002 | 0.09 ± 0.01 | 10.4 | 0.039 ± 0.005 | 0.28 ± 0.01 | 7.1 |
| D753A | 0.064 ± 0.010 | 1.71 ± 0.13 | 26.8 | 0.007 ± 4.0 • 10−4 | 0.14 ± 2.9 • 10−3 | 19.4 | 0.002 ± 2.0 • 10−4 | 0.07 ± 2.4 • 10−3 | 41.2 |
| F755A | 0.135 ± 0.011 | 5.18 ± 0.17 | 38.3 | 0.004 ± 0.001 | 0.07 ± 2.4 • 10−3 | 14.8 | 0.031 ± 0.004 | 0.28 ± 0.01 | 9.0 |
| 0.199 ± 0.015 | 2.90 ± 0.11 | 14.6 | 0.022 ± 0.005 | 0.34 ± 0.03 | 15.6 | 0.027 ± 0.004 | 0.06 ± 3.5 • 10−3 | 2.3 | |
| D753N | 0.121 ± 0.026 | 5.60 ± 0.59 | 46.2 | 0.008 ± 0.001 | 0.13 ± 0.01 | 16.9 | 0.005 ± 3.0 • 10−4 | 0.15 ± 2.4 • 10−3 | 31.1 |
ATPase reactions contained 50 mM HEPES, 100 mM KCl, 20 mM MgSO4, 40 mM NaHCO3, 10 mM ornithine, 1 mM DTT, 10 mM phosphoenolpyruvate, 0.2 mM NADH, 18 u/mL pyruvate kinase, and 24 u/mL lactate dehydrogenase in the presence or absence of 10 mM glutamine. ATP synthesis reactions included 50 mM HEPES, 100 mM KCl, 20 mM MgSO4, 10 mM CP, 10 mM ornithine, 1 mM NADP, 20 mM glucose, 10 u/mL hexokinase, and 5 u/mL glucose-6-phosphate dehydrogenase. All reactions were performed with 5 or 50 μg CPS in 500 μL final volume at 25°C, final pH 7.6. Standard error of the kinetic parameters was determined from nonlinear regression curve fitting (GraFit, version 5.01) and was within ±6.1%.
