Figure 6.
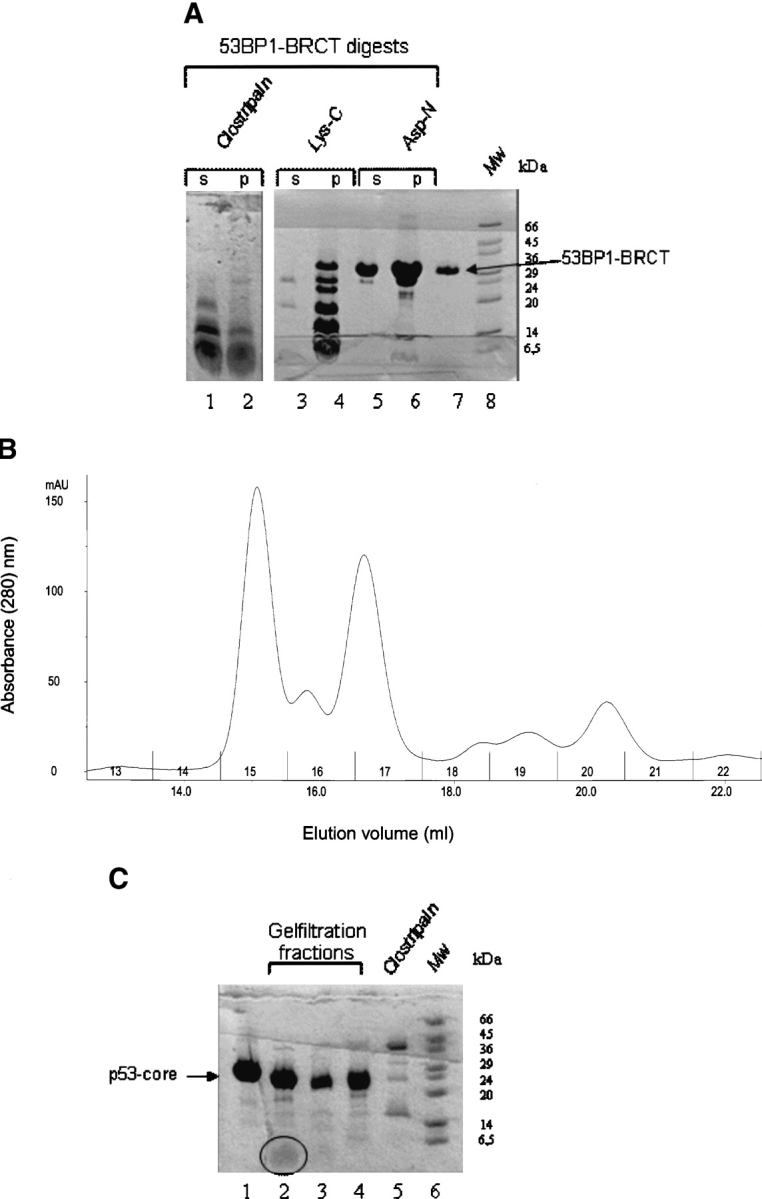
Proteolysis approach for mapping the p53-binding site on 53BP1-BRCT. (A) Digestion of 53BP1-BRCT with proteases clostripain, Lys-C, and Asp-N. Some precipitation occurred during the digestion: ‘s’ and ‘p’ refer to the soluble and the insoluble fractions, respectively, after centrifugation. Only the soluble fraction was used subsequently for binding to p53-core. Intact 53BP1-BRCT is shown in lane 7. Molecular weight markers (Mw) are shown. (B) Analytical gel filtration elution profile of clostripain-digested 53BP1-BRCT incubated with p53-core; collected fractions are numbered. (C) Concentrated fractions from the gel filtration run shown in B. Lane 1 indicates p53-core alone; lane 2, fraction 15; lane 3, fraction 16; and lane 4, fraction 17. The peptide fragment in lane 2 is highlighted by a circle. Fractions 15, 16, and 17 were analyzed by MALDI-TOF mass spectrometry. Comparison of the obtained masses with the masses of expected fragments from clostripain cleavage of 53BP1-BRCT identified the peptides NYLLPAGYSLEEQR (residues 1845–1858) and ILDWQPR (1859–1865) in fraction 15. A peptide corresponding to both these fragments (NYLLPAGYSLEEQRILDWQPR) was identified in fraction 17. A peptide that could correspond to residues 1812–1864 was identified in fraction 16. From the gel, it is clear that there is also some digestion of p53-core by residual clostripain. However, this species is likely to have only small truncations from the termini that should not disrupt the integrity of the structure or the ability to bind 53BP1-BRCT. The shift in elution volume for fraction 15 is larger than expected if the peptides observed by MALDI-TOF bind as monomers, indicating that peptide oligomerization has occurred.
