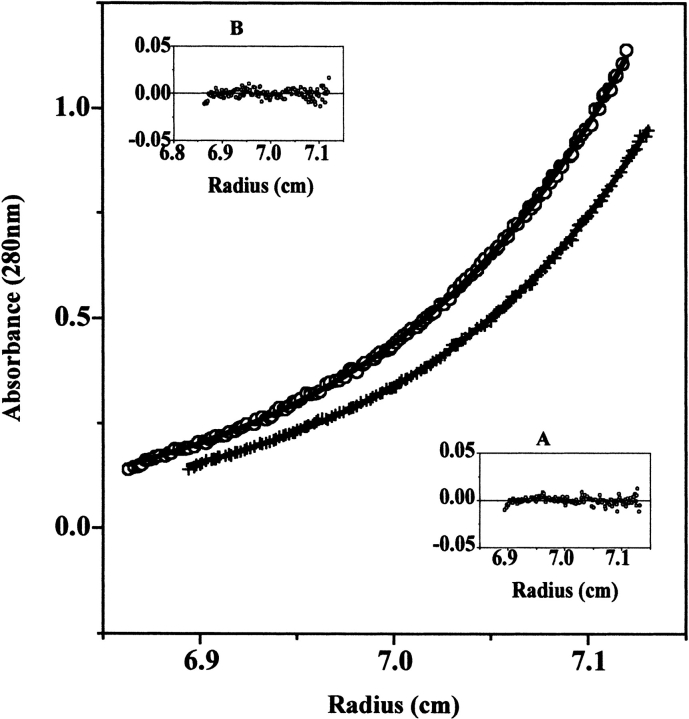
Figure 5.
DMSO effect on the dimerization equilibrium of NAD+ synthetase measured by sedimentation equilibrium at 20°C and 12500 rpm. (Bottom curve) 0.75 mg/mL protein in 60 mM EPPS (pH 8.5) with 20 mM KCl, 19 mM NH4Cl, 10 mM MgCl2, without DMSO. The curve defined by “+” symbols is a mathematical fit using a single ideal species model. (Inset A) Residual plot of the fit. The calculated molecular weight of the species is 61.1 ± 1.4 kD (average of 6 runs), which corresponds to the molecular weight of the NAD+ Synthetase homodimer. (Top curve) 0.75 mg/mL protein in 60 mM EPPS (pH 8.5) with 20 mM KCl, 19 mM NH4Cl, 10 mM MgCl2, 2.5% (v/v) DMSO. The curve defined by open circle symbols is a mathematical global fit to all 6 runs performed at 20°C, using the self-association model. (Inset B) Residual plot of the fit. The calculated association constant (Ka) for dimerization is (5.6 ± 0.02) × 106/M.