Figure 2.
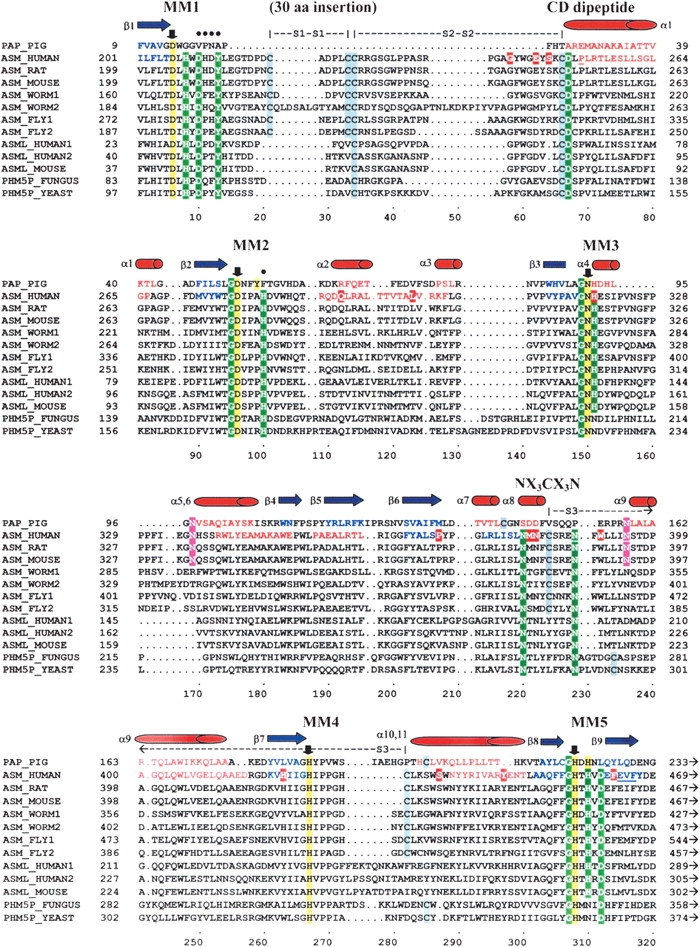
Sequence alignment of the PAP, ASM, and ASM-like phosphoesterase domains. Pig PAP was used as the structural template to build the ASM model. The metal-coordinating motifs 1 through 5 are labeled MM1 through MM5. Down arrows (↓) and yellow highlighted residues indicate known or predicted metal-coordinating residues that are conserved within MM1 to MM5 for the PAP and ASM protein families, respectively (Asp 206, Asp 278, Asn 318, His 425, and His 457 for human ASM). The Cys 250–Asp 251 conserved dipeptide sequence is labeled “CD peptide.” The predicted substrate recognition loop motif is labeled “NX3CX3N.” The residues included in the conserved hydrophilic/aromatic cluster in ASM are marked with solid circles (•). The three disulfide bonds are indicated, respectively, as “S1–S1,” “S2–S2,” and “S3–S3.” The 30-residue insertion (218–247) that could not be modeled is designated. The underlined residues indicate putative misfolded regions of the SMase model, based on an average window length of 10 residues. Highlighted regions indicate the following: Green indicates highly conserved residues in the ASM and ASM-like proteins that are residues often found in DMP active site structures; gray indicates cysteine residues; red indicates known Niemann-Pick mutations in human ASM; magenta indicates predicted N-glycosylation sites for the human and mouse ASM. Red residues and barrels (α1 through α11) are known and predicted α-helices in the PAP and human ASM, respectively. Blue residues and arrows (β1 through β9) are known and predicted β sheets of the PAP and human ASM, respectively. Sequence accession numbers: PAP_PIG–P09889, ASM_HUMAN–P17405, ASM_RAT–XP_219098, ASM_MOUSE–NP_035551, ASM_WORM1–AAC46756, ASML_WORM2–NP_509894, ASML_FLY1–NP_611904, ASML_FLY2–AAF57045, ASML_HUMAN1–NP_055289, ASML_HUMAN2–AH18999, ASML_MOUSE–NP_065586, PHM5P_FUNGUS–T50959, PHM5P_YEAST–NP_010740.
