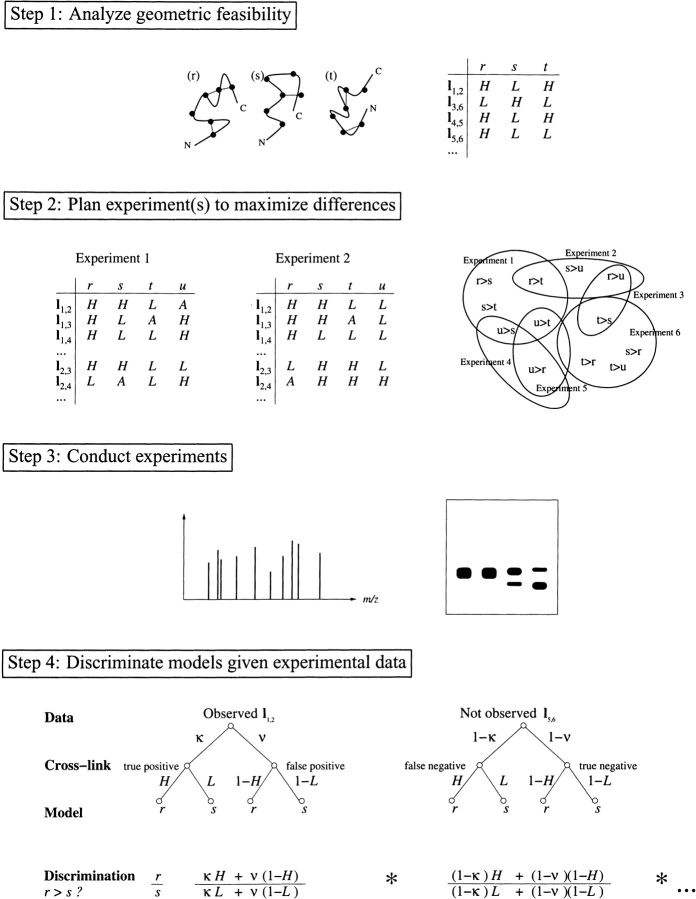
Figure 1.
Model discrimination by cross-linking. (1) Different predicted models of a protein have different patterns of feasible cross-links (dotted lines). Cross-link maps capture the feasibilities (H, L, or A) in terms of conditional relationships for cross-links (rows) given models (columns). (2) Different experimental choices (cross-linkers, mutations) yield different cross-link maps. An experiment could enable selection of one model, if correct, over another (e.g., r > s) if the first model has enough potential positive support (H entries in the cross-link map) where the other doesn’t (L entries). Some experiments provide only ambiguous information for a particular model (A entries). An experiment plan evaluates the relative potential for positive support to select a set of experiments (ovals) to cover the various possible pairwise discriminations. (3) Experimental data, for example, from mass spectrometry or gel electrophoresis, provide support for particular cross-links. (4) Experimental identification of cross-link l1,2 provides evidence for and against models r and s, based on consistency with cross-link maps and modulated by the capture and noise rates of the experimental method (here constant values κ and ν, for capture and noise, respectively). The discrimination ratio combines terms for both observed and unobserved cross-links.