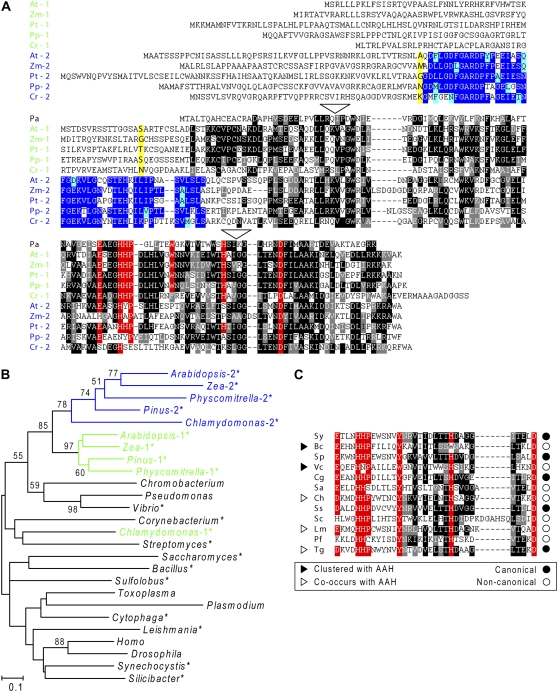
Figure 3.
Primary structures and phylogeny of COG2154 proteins. A, Multiple sequence alignment of representative plant type 1 and type 2 COG2154 proteins. At, Arabidopsis thaliana; Zm, Zea mays; Pt, Pinus taeda; Pp, Physcomitrella patens; Cr, Chlamydomonas reinhardtii. P. aeruginosa (Pa) PCD is included for comparison and defines the approximate extent of the core COG2154 domain common to all sequences in the alignment. The domain unique to type 2 proteins is in blue. Identical residues are shaded in black or dark blue, similar residues in gray or light blue. Residues of the catalytic motif [DE]-x(3)-H-H-P-x(5)-[YW]-x(9)-H-x(8)-D are shaded in red. Residues bolded and shaded in yellow were changed to an initiation codon in the constructs used in complementation assays. Dashes are gaps introduced to maximize alignment. White triangles show positions of two introns present in type 1 and type 2 proteins from Arabidopsis and Physcomitrella and the type 2 protein from Chlamydomonas, but not in other eukaryotic proteins. B, Unrooted neighbor-joining tree for COG2154 proteins from plants and representative species of animals, protists, fungi, and prokaryotes. The sequences analyzed were those tested in this study (asterisked) and those known or inferred from the literature to have PCD activity. Bootstrap values are indicated only for nodes with >50% support. Evolutionary distances are in units of the number of amino acid substitutions per site. C, Sequence alignment of the catalytic motif region of selected non-plant COG2154 proteins from the phylogenetic analysis. Shading is as in part A. Sy, Synechocystis sp.; Bc, Bacillus cereus; Sp, Silicibacter pomeroyi; Vc, Vibrio cholerae; Cg, Corynebacterium glutamicum; Sa, Streptomyces avermitilis; Ch, Cytophaga hutchinsonii; Ss, Sulfolobus solfataricus; Sc, Saccharomyces cerevisiae; Lm, Leishmania major; Pf, Plasmodium falciparum; Tg, Toxoplasma gondii.