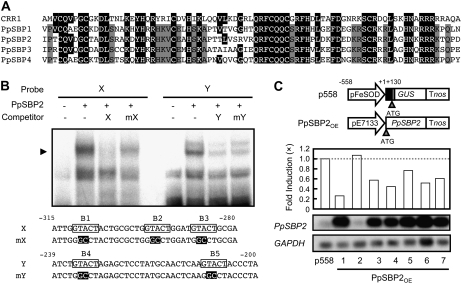
Figure 5.
Binding of transcription factor PpSBP2 to the GTACT motif. A, Alignment of amino acid sequences of the SBP domain of CRR1 and PpSBPs. The highlighted residues indicate amino acids that are identical to those of CRR1. The gray residues indicate amino acids that are identical among PpSBPs. B, Gel retardation assay with in vitro-translated PpSBP2. The specific PpSBP2-DNA complexes are indicated by an arrowhead. Oligonucleotides containing the GTACT motif were used as the probes. The GTACT motif is boxed, and the mutated bases are highlighted. +, Present; −, absent. C, The overexpression of PpSBP2 repressed the expression of the FeSOD gene. Top, Schematic diagram of constructs. The p558 is the reporter construct containing −558 to +130 of the FeSOD gene. The effector construct PpSBP2OE expresses PpSBP2 cDNA under the control of the constitutive pE7133 promoter. Middle, Effect of overexpression PpSBP2 on GUS activities of p558. The bars represent the GUS activities of transgenic moss plants cultured without CuSO4. The GUS activities of the mother transgenic lines containing only the reporter construct were arbitrarily set to 1.0 (dotted line). Bottom, RT-PCR analysis of PpSBP2 expression in transgenic moss plants. After amplification with specific primers, the products were detected by DNA gel-blot hybridization. GAPDH was used as an internal control of RT-PCR.