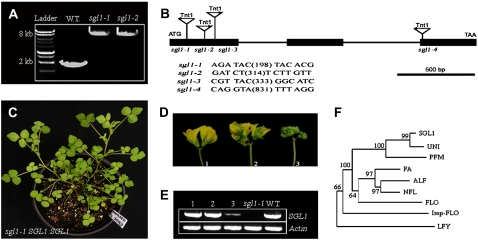
Figure 4.
Molecular cloning of SGL1 gene and functional complementation of sgl1-1 mutant. A, PCR amplification of SGL1gene from wild-type M. truncatula (ecotype R108), sgl1-1, and sgl1-2 mutants. A single insertion of tobacco Tnt1 retrotransposon was detected as a shift in molecular weights of the amplified SGL1gene from each sgl1 mutant allele. B, The intron and exon structure of SGL1 and positions and orientation of Tnt1 insertions in sgl1 mutants. Tnt1 was inserted in the first exon at positions 198 bp, 314 bp, and 333 bp, and in the third exon at the position 831 bp downstream from the translation initiation codon of SGL1 in sgl1-1 to sgl1-4 mutants, respectively. C, A representative sgl1-1 transgenic line transformed with the SGL1 genomic sequence (SGL1:SGL1) exhibited completely rescued wild type-like leaves. D, Representative flowers of three independent sgl1 SGL1:SGL1 transgenic lines, showing completely rescued (line 1), partially rescued (line 2), and nonrescued (line 3) flowers, albeit leaves from all three lines were rescued. E, RT-PCR analysis of SGL1 expression level in developing flowers of three independent sgl1 SGL1:SGL1 transgenic lines (lanes 1–3) compared with that of the sgl1-1 mutant (lane 4) and wild type (lane 5). An Actin gene was used as an internal control (bottom). F, Phylogenetic analysis of SGL1 and its putative orthologs: FLO of snapdragon, LFY of Arabidopsis, NFL of tobacco, UNI of pea, ALF of petunia, Imp-FLO of impatiens, FA of tomato, and PFM of L. japonicus. Bootstrap supports above 50% from 1,000 replicates were shown. [See online article for color version of this figure.]