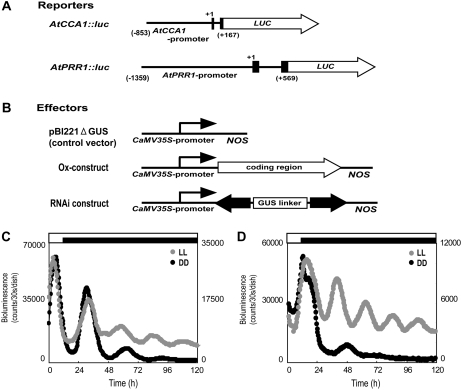
Figure 1.
The semitransient bioluminescence reporter monitoring system. A, Schemes of the reporter constructs. Structures of the AtCCA1 and AtPRR1 promoter regions that drove the firefly luciferase gene (LUC) are shown. Both reporters were constructed as translational fusion genes. Black boxes denote exons of coding regions, and +1 denotes the first base of start codon. (Nakamichi et al., 2004). B, Schemes of the effector constructs. The overexpression effector construct (Ox-construct) and RNAi-mediated knockdown construct (RNAi construct) were derived from the pBI221 vector, in which the coding region or the RNAi construct was under the control of the CaMV 35S promoter and the NOS terminator (Miwa et al., 2006). The cDNA regions used for the RNAi constructs and Ox-constructs for each gene are shown in Supplemental Figure S1. For the control experiments, we used a control vector without any insertion (pBI221ΔGUS). C, Rhythmic expression of bioluminescence following the introduction of the AtCCA1∷luc reporter into L. gibba. Plants that were cultured under 12-h-light/12-h-dark conditions were subjected to particle bombardment. They were treated with an additional 12-h-light/12-h-dark entrainment cycle and then were transferred to a bioluminescence monitoring machine under the experimental light conditions. Bioluminescence profiles of the plants in LL (gray circles) or DD (black circles) are shown (Miwa et al., 2006). The time since the last 12-h dark period is indicated. D, Rhythmic expression of bioluminescence following the introduction of the AtPRR1∷luc reporter. Measurements were performed as in C.